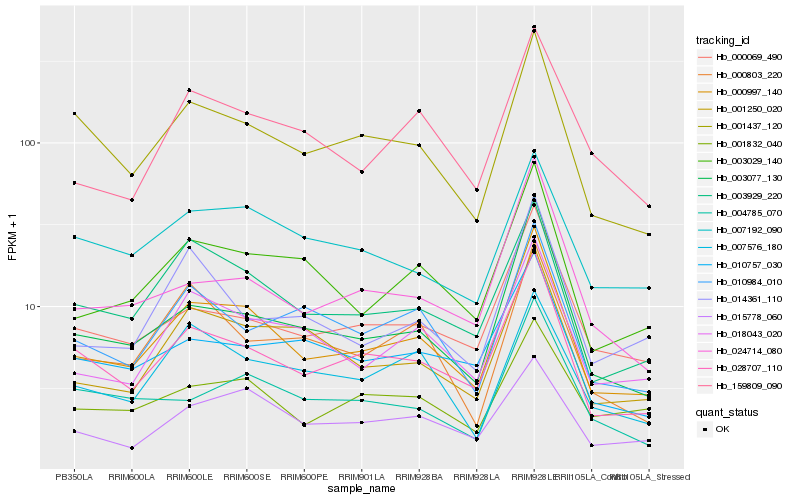
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_140 |
0.0 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_028707_110 |
0.0814509565 |
- |
- |
PREDICTED: uncharacterized protein LOC105645798 [Jatropha curcas] |
| 3 |
Hb_003077_130 |
0.0946753063 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 4 |
Hb_003029_140 |
0.09563464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003929_220 |
0.0980183848 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_018043_020 |
0.0987449022 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001250_020 |
0.1025705607 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000069_490 |
0.1064006412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001437_120 |
0.1075516993 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 10 |
Hb_024714_080 |
0.1154008156 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 11 |
Hb_015778_060 |
0.1155710681 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 12 |
Hb_159809_090 |
0.1159801597 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 13 |
Hb_010984_010 |
0.1206270568 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001832_040 |
0.1220068765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_010757_030 |
0.1227575958 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_014361_110 |
0.127690388 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_004785_070 |
0.1285843513 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000803_220 |
0.1301975982 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 19 |
Hb_007576_180 |
0.1302666566 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 20 |
Hb_007192_090 |
0.1324070945 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |