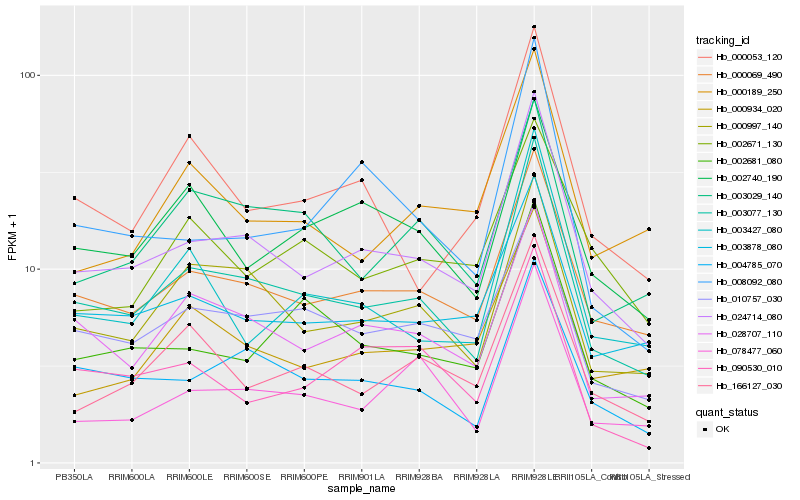
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024714_080 |
0.0 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003077_130 |
0.0663617748 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 3 |
Hb_000069_490 |
0.0767989759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004785_070 |
0.104810541 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_003878_080 |
0.109377363 |
- |
- |
PREDICTED: uncharacterized protein LOC105637045 [Jatropha curcas] |
| 6 |
Hb_028707_110 |
0.1149796744 |
- |
- |
PREDICTED: uncharacterized protein LOC105645798 [Jatropha curcas] |
| 7 |
Hb_000997_140 |
0.1154008156 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_010757_030 |
0.1249529642 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000934_020 |
0.128085032 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 10 |
Hb_008092_080 |
0.1306472315 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 11 |
Hb_002681_080 |
0.1330147005 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000053_120 |
0.1348171165 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_078477_060 |
0.135285071 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 14 |
Hb_166127_030 |
0.1354648189 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_003427_080 |
0.1388066002 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_090530_010 |
0.1417419129 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_002740_190 |
0.1432940472 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000189_250 |
0.1451846941 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003029_140 |
0.1530363788 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002671_130 |
0.1535174486 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |