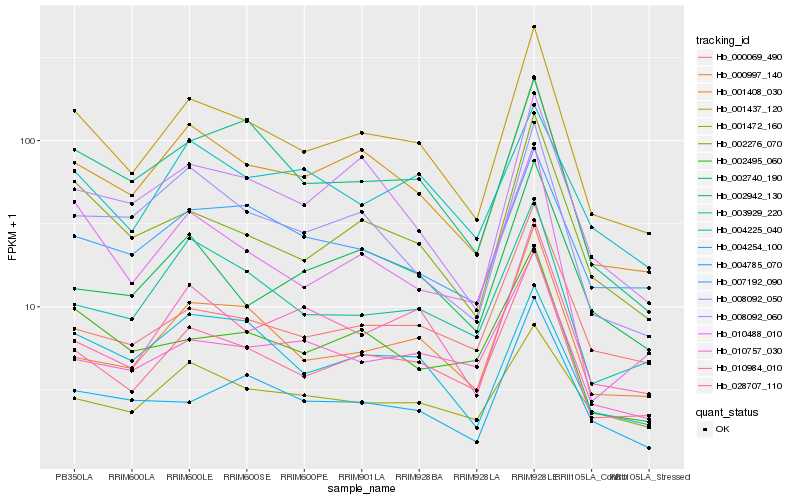
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_120 |
0.0 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 2 |
Hb_028707_110 |
0.0669853788 |
- |
- |
PREDICTED: uncharacterized protein LOC105645798 [Jatropha curcas] |
| 3 |
Hb_001408_030 |
0.077009911 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002276_070 |
0.0929738395 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 5 |
Hb_008092_060 |
0.1051481895 |
- |
- |
- |
| 6 |
Hb_000997_140 |
0.1075516993 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003929_220 |
0.1100721525 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_002942_130 |
0.1103287881 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 9 |
Hb_010488_010 |
0.1110622138 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_008092_050 |
0.1142820318 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 11 |
Hb_007192_090 |
0.1150832368 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 12 |
Hb_002495_060 |
0.1163791946 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_010984_010 |
0.1165491453 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_004254_100 |
0.1192354535 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_010757_030 |
0.1223267944 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_004785_070 |
0.1242681871 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001472_160 |
0.1245667064 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 18 |
Hb_002740_190 |
0.1261440929 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000069_490 |
0.1271942325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004225_040 |
0.1278127504 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |