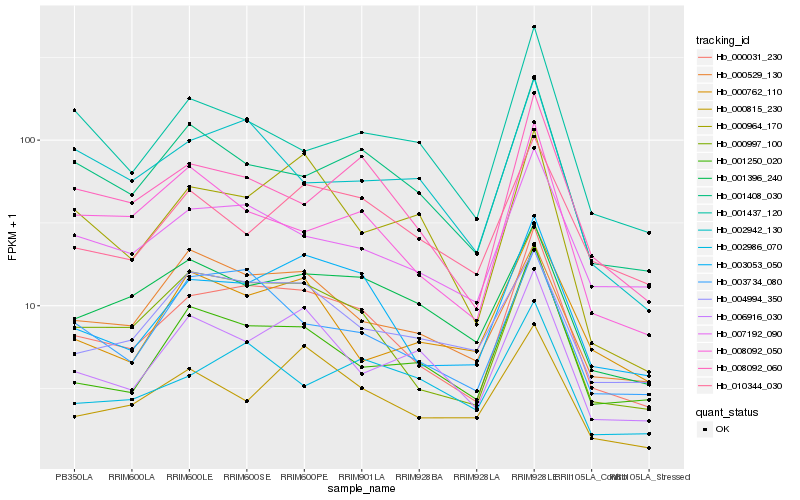
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_230 |
0.0 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003053_050 |
0.0897243284 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 3 |
Hb_000997_100 |
0.1019716211 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X2 [Jatropha curcas] |
| 4 |
Hb_007192_090 |
0.1178272308 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 5 |
Hb_008092_060 |
0.1198707646 |
- |
- |
- |
| 6 |
Hb_001250_020 |
0.1228532484 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001408_030 |
0.1254549701 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003734_080 |
0.1286045722 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 9 |
Hb_006916_030 |
0.1334425777 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 10 |
Hb_008092_050 |
0.1361943223 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 11 |
Hb_001396_240 |
0.136236965 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004994_350 |
0.1395596512 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002986_070 |
0.1404679419 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 [Jatropha curcas] |
| 14 |
Hb_002942_130 |
0.1409573398 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 15 |
Hb_010344_030 |
0.1441720631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001437_120 |
0.1441781066 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 17 |
Hb_000529_130 |
0.1455269516 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 18 |
Hb_000815_230 |
0.1458379933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000964_170 |
0.1466121462 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 20 |
Hb_000762_110 |
0.1476817585 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |