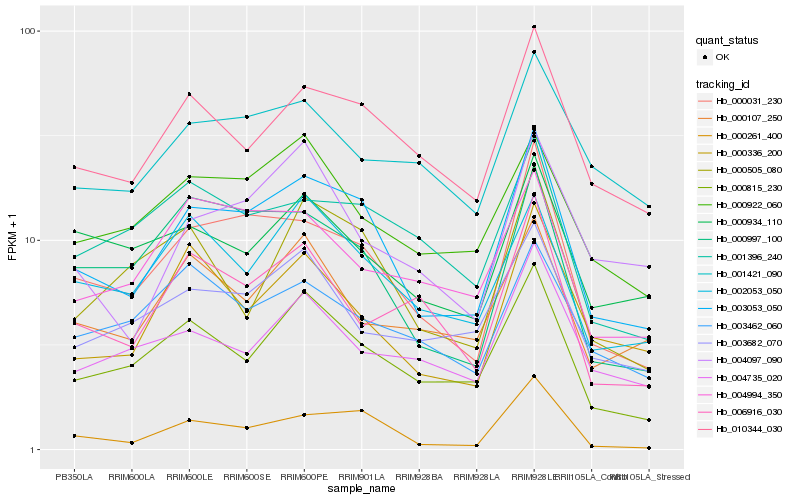
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003053_050 |
0.0 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 2 |
Hb_000031_230 |
0.0897243284 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_010344_030 |
0.1121917336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000815_230 |
0.1129403349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000997_100 |
0.1301817644 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002053_050 |
0.1338692821 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 7 |
Hb_000336_200 |
0.1377452002 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 8 |
Hb_000261_400 |
0.1392672992 |
- |
- |
hypothetical protein JCGZ_01125 [Jatropha curcas] |
| 9 |
Hb_000922_060 |
0.142154824 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 10 |
Hb_000505_080 |
0.1431367686 |
- |
- |
Inner membrane transport protein yjjL, putative [Ricinus communis] |
| 11 |
Hb_003682_070 |
0.1452162159 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001396_240 |
0.1453525382 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004994_350 |
0.1474861858 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000934_110 |
0.1484237143 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |
| 15 |
Hb_000107_250 |
0.1485663928 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 16 |
Hb_004097_090 |
0.1492417077 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 17 |
Hb_004735_020 |
0.1499412401 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 18 |
Hb_001421_090 |
0.1508704278 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 19 |
Hb_006916_030 |
0.1511550795 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 20 |
Hb_003462_060 |
0.1516945912 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |