| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
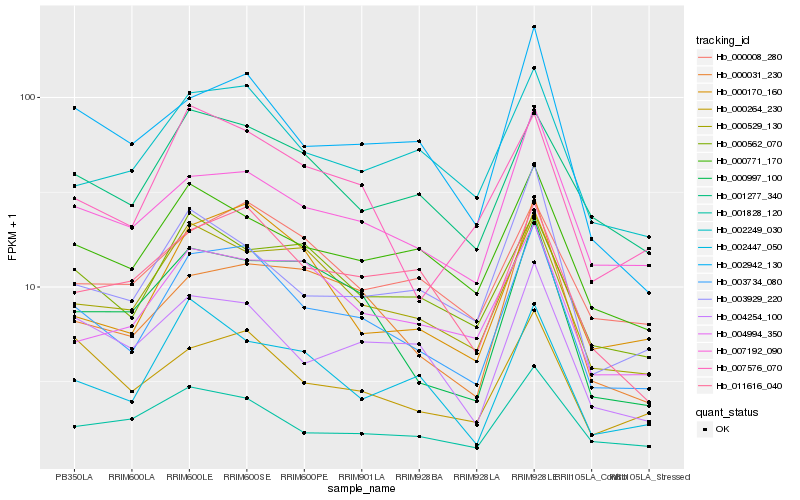
Hb_003734_080 |
0.0 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 2 |
Hb_002249_030 |
0.109532801 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007192_090 |
0.1115579775 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 4 |
Hb_002942_130 |
0.1119673522 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004254_100 |
0.1158563649 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000264_230 |
0.1170861656 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 7 |
Hb_000170_160 |
0.117110646 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000529_130 |
0.117327579 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 9 |
Hb_011616_040 |
0.1173999454 |
- |
- |
glucan water dikinase 3, partial [Manihot esculenta] |
| 10 |
Hb_001277_340 |
0.1181963599 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 11 |
Hb_000771_170 |
0.1188215682 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_007576_070 |
0.1237122597 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 13 |
Hb_000997_100 |
0.1238867741 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001828_120 |
0.1249078855 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 15 |
Hb_003929_220 |
0.1273201599 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000008_280 |
0.1278442548 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 17 |
Hb_000031_230 |
0.1286045722 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002447_050 |
0.1292641432 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000562_070 |
0.1294183244 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004994_350 |
0.1295774075 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |