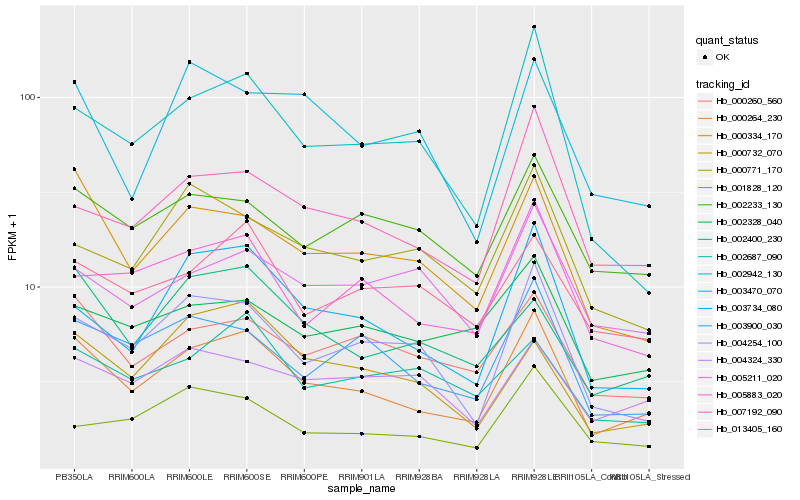
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649528 [Jatropha curcas] |
| 2 |
Hb_000334_170 |
0.1105503577 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003900_030 |
0.1151084195 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41720 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003734_080 |
0.1170861656 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 5 |
Hb_000260_560 |
0.1190705055 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007192_090 |
0.1195452118 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 7 |
Hb_004254_100 |
0.1197758148 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005883_020 |
0.1220241409 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74850, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002942_130 |
0.1264110845 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002233_130 |
0.1275509239 |
- |
- |
transporter, putative [Ricinus communis] |
| 11 |
Hb_002400_230 |
0.1332251499 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 12 |
Hb_013405_160 |
0.134990378 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002328_040 |
0.1363847211 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001828_120 |
0.1397982488 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 15 |
Hb_000732_070 |
0.1432241777 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 16 |
Hb_004324_330 |
0.1435854188 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 17 |
Hb_000771_170 |
0.1441594279 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_002687_090 |
0.1446649058 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_003470_070 |
0.1450957859 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 20 |
Hb_005211_020 |
0.1453324935 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |