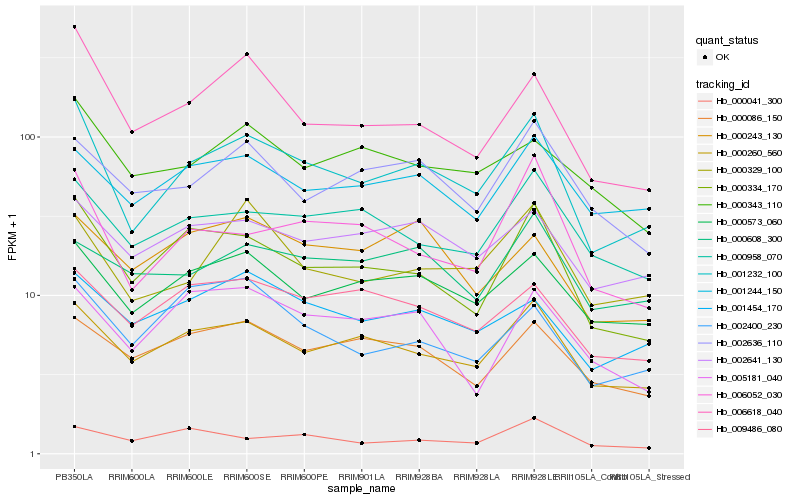
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001232_100 |
0.0 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 2 |
Hb_000260_560 |
0.1068299755 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000334_170 |
0.1069347207 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006618_040 |
0.1166931467 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 5 |
Hb_000573_060 |
0.1197039967 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 6 |
Hb_006052_030 |
0.1242236204 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A [Jatropha curcas] |
| 7 |
Hb_000329_100 |
0.1292845389 |
- |
- |
PREDICTED: uncharacterized protein LOC105643144 [Jatropha curcas] |
| 8 |
Hb_000958_070 |
0.132381491 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 9 |
Hb_002641_130 |
0.1330620192 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 10 |
Hb_001244_150 |
0.1394653173 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 11 |
Hb_009486_080 |
0.1399011458 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 12 |
Hb_001454_170 |
0.1415597528 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 13 |
Hb_000343_110 |
0.1423623337 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 14 |
Hb_002636_110 |
0.1431057782 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_000086_150 |
0.1453652005 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002400_230 |
0.1511118227 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 17 |
Hb_000243_130 |
0.1528411178 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 18 |
Hb_000041_300 |
0.1538666253 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized ATP-dependent helicase C23E6.02 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000608_300 |
0.1542612208 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 20 |
Hb_005181_040 |
0.1544654331 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |