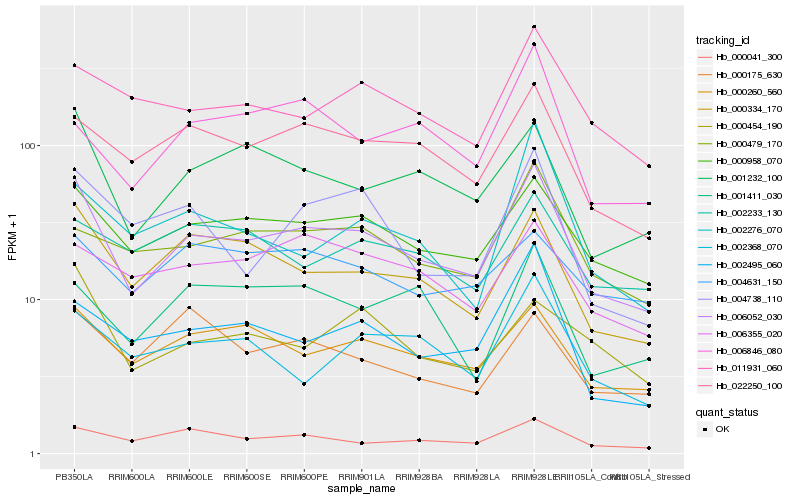
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006052_030 |
0.0 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A [Jatropha curcas] |
| 2 |
Hb_000958_070 |
0.1095013794 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 3 |
Hb_000334_170 |
0.1212636774 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001232_100 |
0.1242236204 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 5 |
Hb_000260_560 |
0.1273282201 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 6 |
Hb_022250_100 |
0.1277661849 |
- |
- |
calnexin, putative [Ricinus communis] |
| 7 |
Hb_000041_300 |
0.1337076737 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized ATP-dependent helicase C23E6.02 isoform X2 [Jatropha curcas] |
| 8 |
Hb_011931_060 |
0.1406197359 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 9 |
Hb_002495_060 |
0.1407721492 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000479_170 |
0.1445264908 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 11 |
Hb_002368_070 |
0.1459712677 |
- |
- |
PREDICTED: uncharacterized protein LOC105634087 [Jatropha curcas] |
| 12 |
Hb_004738_110 |
0.1472135655 |
- |
- |
phosphoenolpyruvate carboxylase [Manihot esculenta] |
| 13 |
Hb_000454_190 |
0.1555730934 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_006355_020 |
0.1563948919 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 15 |
Hb_006846_080 |
0.1590201494 |
- |
- |
calnexin, putative [Ricinus communis] |
| 16 |
Hb_001411_030 |
0.1600762319 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 17 |
Hb_002233_130 |
0.1611915401 |
- |
- |
transporter, putative [Ricinus communis] |
| 18 |
Hb_002276_070 |
0.1631533332 |
- |
- |
PREDICTED: heat shock protein 83 [Jatropha curcas] |
| 19 |
Hb_000175_630 |
0.1644645281 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g11310, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004631_150 |
0.1645846913 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |