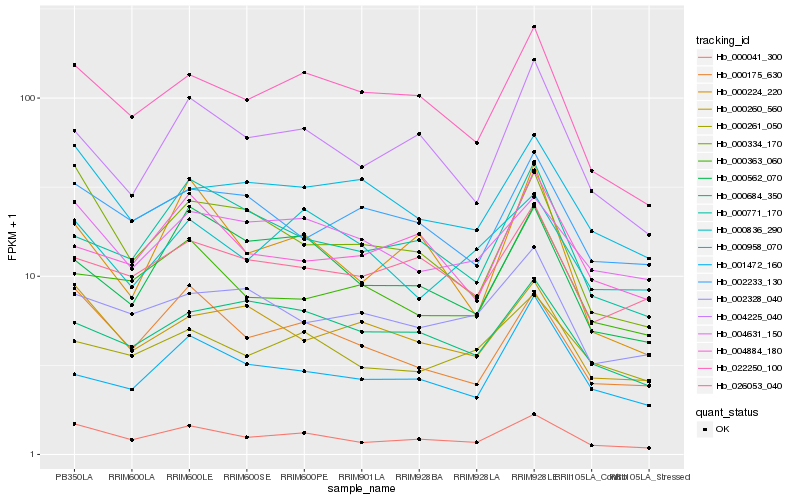
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_300 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized ATP-dependent helicase C23E6.02 isoform X2 [Jatropha curcas] |
| 2 |
Hb_022250_100 |
0.0869188407 |
- |
- |
calnexin, putative [Ricinus communis] |
| 3 |
Hb_000175_630 |
0.0940014904 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g11310, mitochondrial [Jatropha curcas] |
| 4 |
Hb_004225_040 |
0.1016264552 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 5 |
Hb_000334_170 |
0.112774343 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 6 |
Hb_026053_040 |
0.1136871049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000261_050 |
0.1149571088 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 8 |
Hb_000958_070 |
0.1155796689 |
- |
- |
hypothetical protein PRUPE_ppa007681mg [Prunus persica] |
| 9 |
Hb_004631_150 |
0.1160347849 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000363_060 |
0.1180294452 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002233_130 |
0.1180690364 |
- |
- |
transporter, putative [Ricinus communis] |
| 12 |
Hb_000260_560 |
0.1203023925 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000771_170 |
0.1206747744 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_002328_040 |
0.1232679211 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000562_070 |
0.1237218193 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000684_350 |
0.1248092228 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004884_180 |
0.1288110862 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000836_290 |
0.1294599813 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_001472_160 |
0.1305441891 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 20 |
Hb_000224_220 |
0.1305672994 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |