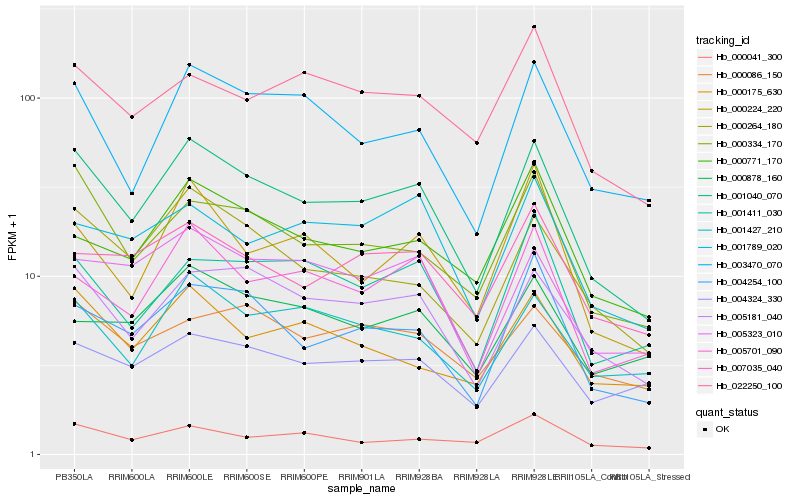
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001040_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649523 [Jatropha curcas] |
| 2 |
Hb_005181_040 |
0.0886242638 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 3 |
Hb_000264_180 |
0.1003208008 |
- |
- |
PREDICTED: uncharacterized protein LOC100777318 [Glycine max] |
| 4 |
Hb_000334_170 |
0.1012952958 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP42 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003470_070 |
0.1044674848 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 6 |
Hb_004254_100 |
0.1049097467 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005701_090 |
0.1090205688 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001427_210 |
0.112150099 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005323_010 |
0.1143679217 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 10 |
Hb_000175_630 |
0.1204285482 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g11310, mitochondrial [Jatropha curcas] |
| 11 |
Hb_007035_040 |
0.1232998519 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004324_330 |
0.1251882891 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 13 |
Hb_001411_030 |
0.1253288552 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 14 |
Hb_000224_220 |
0.1260659181 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |
| 15 |
Hb_000771_170 |
0.1261506304 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_000086_150 |
0.1267964284 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_022250_100 |
0.1295018738 |
- |
- |
calnexin, putative [Ricinus communis] |
| 18 |
Hb_001789_020 |
0.1298905654 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000041_300 |
0.1327478681 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized ATP-dependent helicase C23E6.02 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000878_160 |
0.1339516238 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |