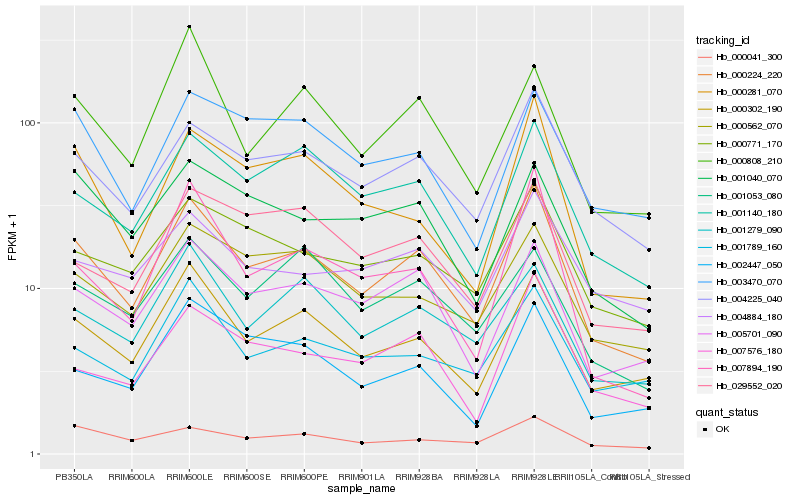
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000224_220 |
0.0 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |
| 2 |
Hb_000302_190 |
0.0873960312 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 3 |
Hb_004225_040 |
0.0885664226 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 4 |
Hb_005701_090 |
0.0990198759 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_001140_180 |
0.1067064881 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 6 |
Hb_000771_170 |
0.113564051 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_001279_090 |
0.1141812487 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 8 |
Hb_001789_160 |
0.1155870457 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 9 |
Hb_007894_190 |
0.1172209131 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 10 |
Hb_000281_070 |
0.1200441519 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 11 |
Hb_000562_070 |
0.1218356732 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000808_210 |
0.1224195999 |
- |
- |
hypothetical protein EUTSA_v10023481mg [Eutrema salsugineum] |
| 13 |
Hb_007576_180 |
0.1233741479 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 14 |
Hb_002447_050 |
0.1238974683 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_003470_070 |
0.1250808661 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 16 |
Hb_029552_020 |
0.1259770654 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001040_070 |
0.1260659181 |
- |
- |
PREDICTED: uncharacterized protein LOC105649523 [Jatropha curcas] |
| 18 |
Hb_004884_180 |
0.1299119659 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000041_300 |
0.1305672994 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized ATP-dependent helicase C23E6.02 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001053_080 |
0.1307530802 |
- |
- |
OsCesA3 protein [Morus notabilis] |