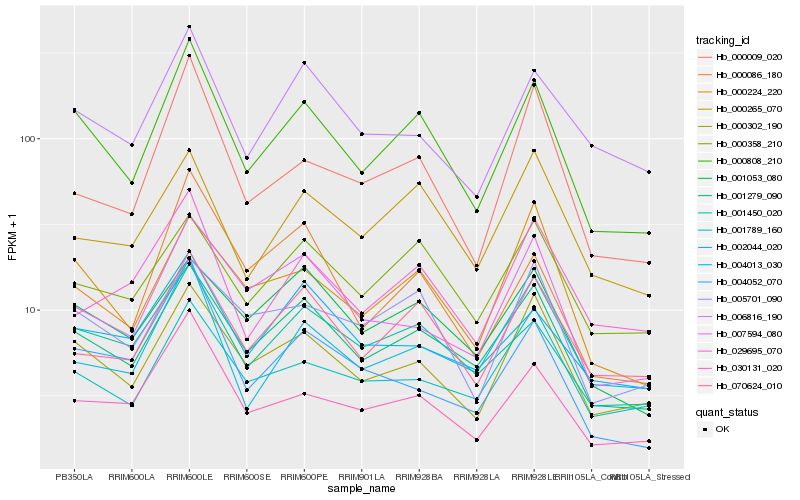
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000808_210 |
0.0 |
- |
- |
hypothetical protein EUTSA_v10023481mg [Eutrema salsugineum] |
| 2 |
Hb_001279_090 |
0.0860589567 |
- |
- |
PREDICTED: uncharacterized protein LOC105633322 [Jatropha curcas] |
| 3 |
Hb_000302_190 |
0.0941420152 |
- |
- |
PREDICTED: uncharacterized protein At3g49140-like [Jatropha curcas] |
| 4 |
Hb_030131_020 |
0.1100887186 |
- |
- |
Carboxypeptidase B2 precursor, putative [Ricinus communis] |
| 5 |
Hb_001450_020 |
0.1119760921 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_070624_010 |
0.1144273506 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 7 |
Hb_000224_220 |
0.1224195999 |
- |
- |
PREDICTED: protein disulfide isomerase-like 2-3 [Jatropha curcas] |
| 8 |
Hb_000358_210 |
0.1271593172 |
- |
- |
oligosaccharyl transferase, putative [Ricinus communis] |
| 9 |
Hb_001053_080 |
0.1273489284 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 10 |
Hb_005701_090 |
0.1274266435 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_006816_190 |
0.1284459143 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 12 |
Hb_000265_070 |
0.1313950313 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 13 |
Hb_000009_020 |
0.1326082088 |
- |
- |
ketol-acid reductoisomerase, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001789_160 |
0.1333698772 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 15 |
Hb_007594_080 |
0.133594224 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 16 |
Hb_000086_180 |
0.1366098927 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |
| 17 |
Hb_002044_020 |
0.1375548992 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 18 |
Hb_029695_070 |
0.1384608785 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_004013_030 |
0.1389609165 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 20 |
Hb_004052_070 |
0.1399231659 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |