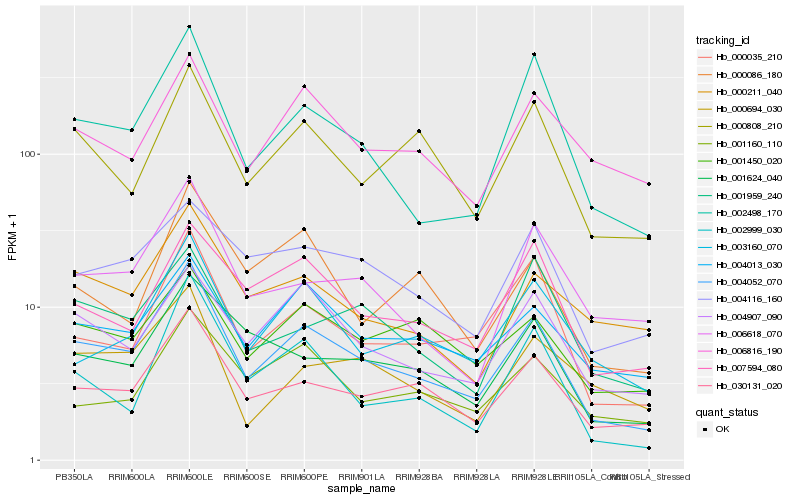
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_070 |
0.0 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 2 |
Hb_030131_020 |
0.1078827386 |
- |
- |
Carboxypeptidase B2 precursor, putative [Ricinus communis] |
| 3 |
Hb_002498_170 |
0.1161512971 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000035_210 |
0.1309427556 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 5 |
Hb_004907_090 |
0.1352279535 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_004013_030 |
0.137292008 |
- |
- |
Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, putative [Ricinus communis] |
| 7 |
Hb_003160_070 |
0.1397035894 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 8 |
Hb_000808_210 |
0.1399231659 |
- |
- |
hypothetical protein EUTSA_v10023481mg [Eutrema salsugineum] |
| 9 |
Hb_002999_030 |
0.1406273198 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 10 |
Hb_001624_040 |
0.1440521207 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007594_080 |
0.1450542565 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 12 |
Hb_006618_070 |
0.1461813171 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001450_020 |
0.1490002524 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000211_040 |
0.1496966464 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 15 |
Hb_006816_190 |
0.1510855198 |
- |
- |
calreticulin family protein [Populus trichocarpa] |
| 16 |
Hb_004116_160 |
0.1515203875 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000694_030 |
0.1536947307 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 18 |
Hb_001160_110 |
0.1539819596 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 19 |
Hb_000086_180 |
0.1541393704 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |
| 20 |
Hb_001959_240 |
0.1546333587 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |