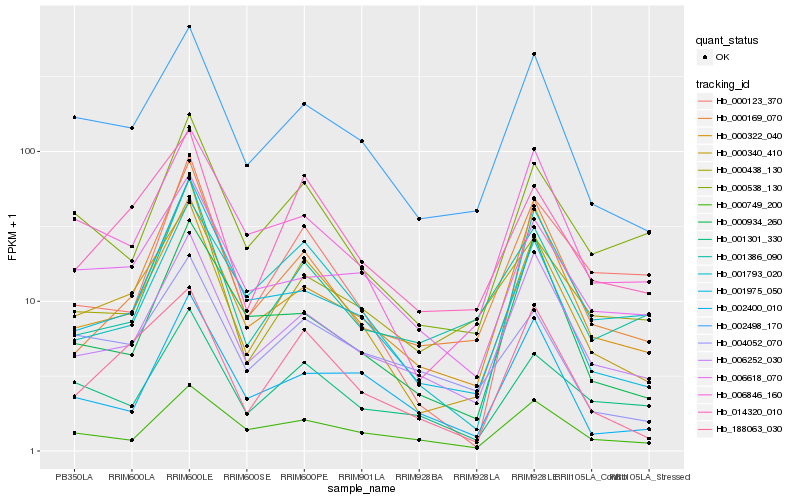
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_410 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 2 |
Hb_002498_170 |
0.0997334785 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_188063_030 |
0.1039254275 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 4 |
Hb_014320_010 |
0.1131099424 |
- |
- |
PREDICTED: uncharacterized protein LOC105629932 [Jatropha curcas] |
| 5 |
Hb_006252_030 |
0.1280201288 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 6 |
Hb_006846_160 |
0.1381177818 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 7 |
Hb_000322_040 |
0.1420423558 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000169_070 |
0.1527020385 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 9 |
Hb_000538_130 |
0.1559242428 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 10 |
Hb_001301_330 |
0.1607819296 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 11 |
Hb_001975_050 |
0.1624741683 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 12 |
Hb_000749_200 |
0.1644285437 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001793_020 |
0.165272839 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 14 |
Hb_006618_070 |
0.16650603 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004052_070 |
0.1682221216 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 16 |
Hb_000934_260 |
0.1709448977 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000438_130 |
0.175528679 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 18 |
Hb_002400_010 |
0.1757557023 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 19 |
Hb_001386_090 |
0.1759371889 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 20 |
Hb_000123_370 |
0.1784677393 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |