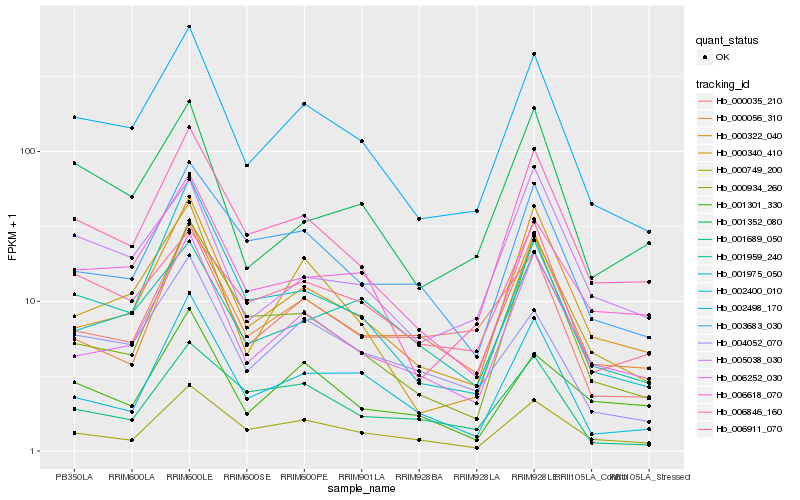
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002498_170 |
0.0 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000340_410 |
0.0997334785 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_006846_160 |
0.1051809033 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 4 |
Hb_006252_030 |
0.1092210805 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 5 |
Hb_004052_070 |
0.1161512971 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 6 |
Hb_006618_070 |
0.1178062458 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000322_040 |
0.1222720743 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000749_200 |
0.1276442289 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001959_240 |
0.1322021913 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit B, chloroplastic/mitochondrial [Jatropha curcas] |
| 10 |
Hb_003683_030 |
0.1327852263 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 11 |
Hb_002400_010 |
0.1331221417 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 12 |
Hb_006911_070 |
0.1335112333 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 13 |
Hb_001352_080 |
0.1335935589 |
- |
- |
PREDICTED: uncharacterized protein At5g02240 [Jatropha curcas] |
| 14 |
Hb_000035_210 |
0.1346558839 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 15 |
Hb_000934_260 |
0.1427342857 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000056_310 |
0.1460195654 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 17 |
Hb_005038_030 |
0.1474963396 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001975_050 |
0.1506443574 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |
| 19 |
Hb_001301_330 |
0.1530280029 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 20 |
Hb_001689_050 |
0.154334226 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |