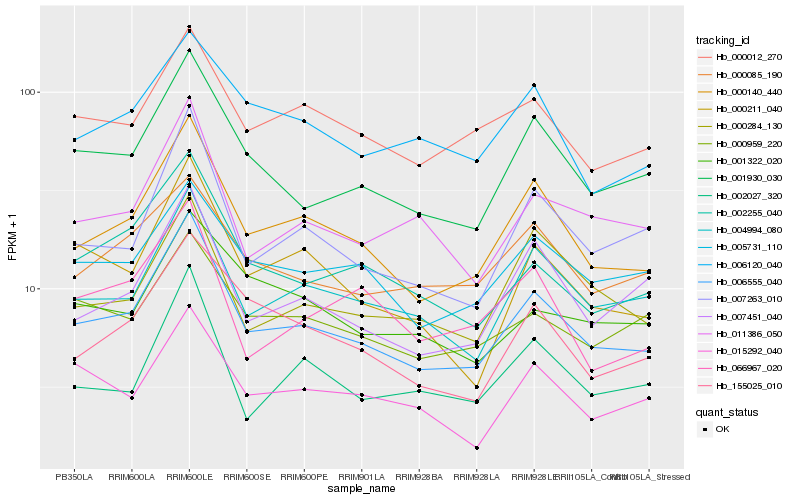
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006555_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 2 |
Hb_000140_440 |
0.0549236776 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 3 |
Hb_001930_030 |
0.0772657968 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004994_080 |
0.0797155604 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_007263_010 |
0.084297904 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 6 |
Hb_002255_040 |
0.0914931129 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 7 |
Hb_011386_050 |
0.0965682574 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_007451_040 |
0.0979354004 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 9 |
Hb_000211_040 |
0.0984594977 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 10 |
Hb_005731_110 |
0.10186181 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 11 |
Hb_000012_270 |
0.1019170007 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 12 |
Hb_155025_010 |
0.1035576447 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_002027_320 |
0.107333526 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 14 |
Hb_015292_040 |
0.1073563549 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 15 |
Hb_000959_220 |
0.1092997848 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000085_190 |
0.1099663527 |
- |
- |
FtsZ1 protein [Manihot esculenta] |
| 17 |
Hb_000284_130 |
0.1111666069 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 18 |
Hb_006120_040 |
0.1127414125 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 19 |
Hb_001322_020 |
0.1156194925 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 20 |
Hb_066967_020 |
0.1161986577 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |