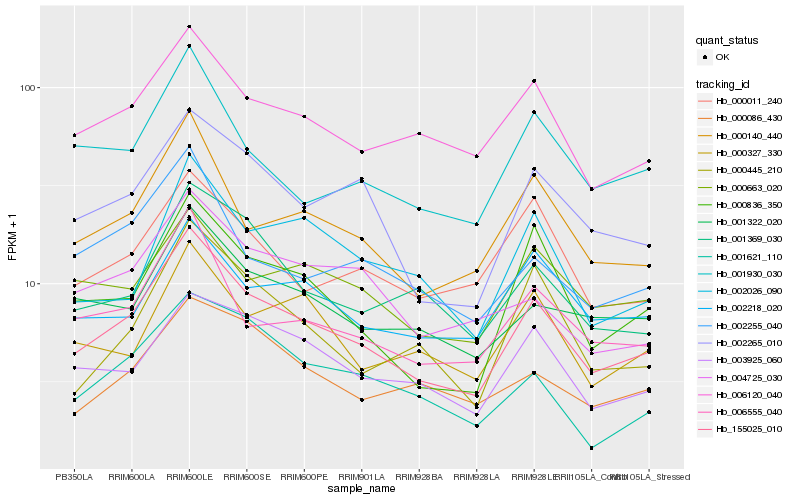
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155025_010 |
0.0 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 2 |
Hb_000140_440 |
0.0927427857 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 3 |
Hb_002265_010 |
0.0990412487 |
- |
- |
ef-hand calcium binding protein, putative [Ricinus communis] |
| 4 |
Hb_006120_040 |
0.1027164476 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 5 |
Hb_006555_040 |
0.1035576447 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 6 |
Hb_000836_350 |
0.1059622724 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001322_020 |
0.1081362252 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 8 |
Hb_003925_060 |
0.1111096671 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001369_030 |
0.1149700794 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 10 |
Hb_001930_030 |
0.1162763259 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002255_040 |
0.1179908504 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 12 |
Hb_002218_020 |
0.1181770068 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 13 |
Hb_000086_430 |
0.1209151422 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 14 |
Hb_000445_210 |
0.121061258 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 15 |
Hb_004725_030 |
0.1222997417 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 16 |
Hb_000327_330 |
0.1224397543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000663_020 |
0.122556327 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_000011_240 |
0.1226949804 |
- |
- |
PREDICTED: uncharacterized protein LOC105631316 [Jatropha curcas] |
| 19 |
Hb_002026_090 |
0.1228623645 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 20 |
Hb_001621_110 |
0.1236400058 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |