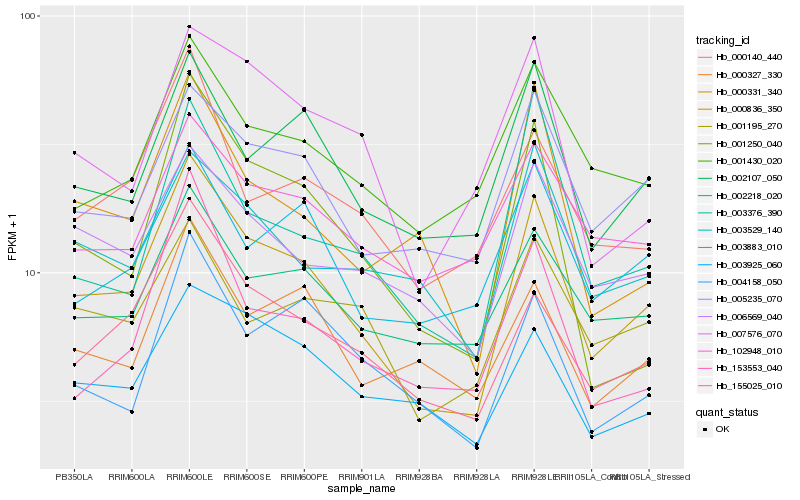
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_350 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 2 |
Hb_155025_010 |
0.1059622724 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 3 |
Hb_005235_070 |
0.1160270169 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002107_050 |
0.1180380155 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 5 |
Hb_006569_040 |
0.1198670817 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 6 |
Hb_000140_440 |
0.1279691982 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 7 |
Hb_002218_020 |
0.1291303764 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 8 |
Hb_003883_010 |
0.1299315607 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 9 |
Hb_000327_330 |
0.1303991083 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004158_050 |
0.1304398658 |
- |
- |
hypothetical protein JCGZ_09026 [Jatropha curcas] |
| 11 |
Hb_003925_060 |
0.1307918077 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000331_340 |
0.1320818148 |
- |
- |
PREDICTED: uncharacterized protein LOC105640234 [Jatropha curcas] |
| 13 |
Hb_001430_020 |
0.1329633664 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_003529_140 |
0.1333825375 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001250_040 |
0.1342656467 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 16 |
Hb_001195_270 |
0.1350639516 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007576_070 |
0.1365310328 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X3 [Jatropha curcas] |
| 18 |
Hb_003376_390 |
0.1375803994 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 19 |
Hb_153553_040 |
0.1381886058 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 20 |
Hb_102948_010 |
0.1383768323 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |