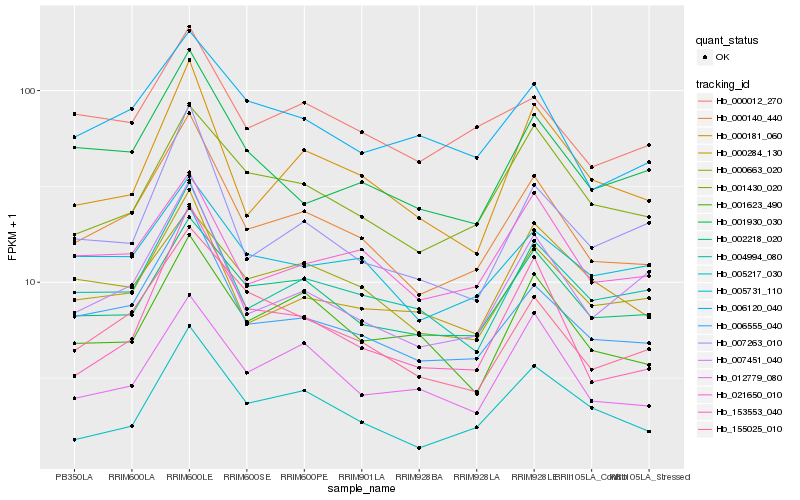
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_440 |
0.0 |
- |
- |
PREDICTED: probable hydroxyacylglutathione hydrolase 2, chloroplast [Jatropha curcas] |
| 2 |
Hb_006555_040 |
0.0549236776 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 3 |
Hb_004994_080 |
0.092215147 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_155025_010 |
0.0927427857 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_007451_040 |
0.0977834315 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 6 |
Hb_002218_020 |
0.1005530382 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 7 |
Hb_000181_060 |
0.1028476036 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 8 |
Hb_007263_010 |
0.1038558046 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_000012_270 |
0.1046300183 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 10 |
Hb_001930_030 |
0.1059110898 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005731_110 |
0.1067626057 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 12 |
Hb_000663_020 |
0.1077771786 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_021650_010 |
0.1079413046 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 14 |
Hb_006120_040 |
0.1088028868 |
- |
- |
PREDICTED: prostatic spermine-binding protein [Jatropha curcas] |
| 15 |
Hb_001430_020 |
0.1114907273 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_005217_030 |
0.1123422266 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 17 |
Hb_001623_490 |
0.1123441838 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 18 |
Hb_153553_040 |
0.1127015512 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 19 |
Hb_000284_130 |
0.1139141324 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 20 |
Hb_012779_080 |
0.1161772455 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |