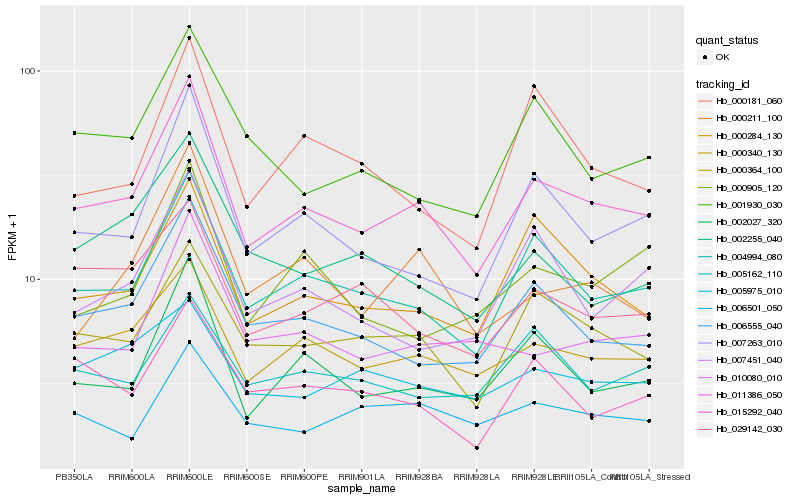
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011386_050 |
0.0 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_004994_080 |
0.0794119722 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000340_130 |
0.0887673054 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002027_320 |
0.0961181635 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 5 |
Hb_006555_040 |
0.0965682574 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 6 |
Hb_007263_010 |
0.0974680418 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 7 |
Hb_000364_100 |
0.0978278668 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 8 |
Hb_000284_130 |
0.1115418695 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 9 |
Hb_010080_010 |
0.1156830004 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 10 |
Hb_000211_100 |
0.1213747644 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 11 |
Hb_029142_030 |
0.1236164735 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 12 |
Hb_001930_030 |
0.1245071632 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 13 |
Hb_015292_040 |
0.1247004986 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 14 |
Hb_002255_040 |
0.1256541119 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 15 |
Hb_007451_040 |
0.1278520735 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 16 |
Hb_006501_050 |
0.131263846 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 17 |
Hb_000905_120 |
0.1313812344 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 18 |
Hb_005975_010 |
0.1314080806 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_005162_110 |
0.1315782779 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 20 |
Hb_000181_060 |
0.1320574491 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |