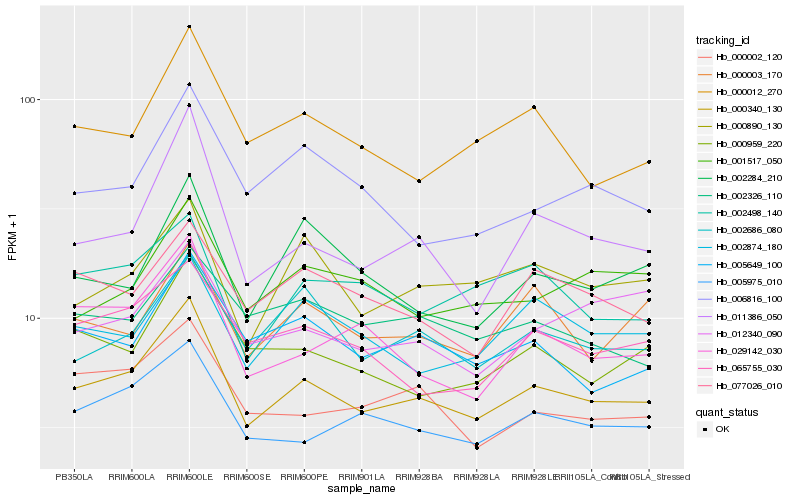
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005975_010 |
0.0859493488 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_011386_050 |
0.0887673054 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_005649_100 |
0.0958171701 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 5 |
Hb_002686_080 |
0.0959397915 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 6 |
Hb_000012_270 |
0.0968702758 |
- |
- |
NADH-plastoquinone oxidoreductase, putative [Ricinus communis] |
| 7 |
Hb_000002_120 |
0.0972110228 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 8 |
Hb_000959_220 |
0.0978666935 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_029142_030 |
0.0988332791 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 10 |
Hb_065755_030 |
0.0990221681 |
- |
- |
- |
| 11 |
Hb_000890_130 |
0.0991227905 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 12 |
Hb_002874_180 |
0.1037259473 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002498_140 |
0.1045837019 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 14 |
Hb_006816_100 |
0.1056374254 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 15 |
Hb_077026_010 |
0.1056499358 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 16 |
Hb_001517_050 |
0.1064064625 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000003_170 |
0.1066501632 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 18 |
Hb_002284_210 |
0.107226988 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 19 |
Hb_002326_110 |
0.1073050455 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 20 |
Hb_012340_090 |
0.1087033707 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |