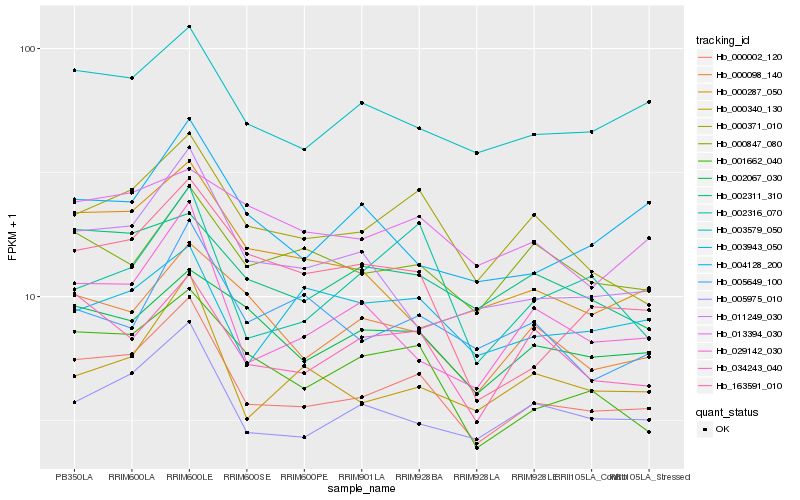
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 2 |
Hb_005975_010 |
0.07944766 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_003579_050 |
0.0841402501 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 4 |
Hb_000371_010 |
0.084761734 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 5 |
Hb_001662_040 |
0.0913320402 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 6 |
Hb_034243_040 |
0.0939633444 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1-like [Jatropha curcas] |
| 7 |
Hb_000098_140 |
0.0967155343 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 8 |
Hb_002067_030 |
0.0970525787 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000340_130 |
0.0972110228 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 10 |
Hb_029142_030 |
0.0976435987 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 11 |
Hb_002311_310 |
0.0988343005 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 12 |
Hb_000847_080 |
0.1054994943 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Jatropha curcas] |
| 13 |
Hb_011249_030 |
0.1085917786 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 14 |
Hb_013394_030 |
0.1094652905 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 15 |
Hb_163591_010 |
0.1109562495 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Populus euphratica] |
| 16 |
Hb_004128_200 |
0.111986249 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 17 |
Hb_003943_050 |
0.1127087883 |
- |
- |
phosphoglucomutase, putative [Ricinus communis] |
| 18 |
Hb_000287_050 |
0.1137589629 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 19 |
Hb_005649_100 |
0.1137860488 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 20 |
Hb_002316_070 |
0.1149177925 |
- |
- |
PREDICTED: 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial [Jatropha curcas] |