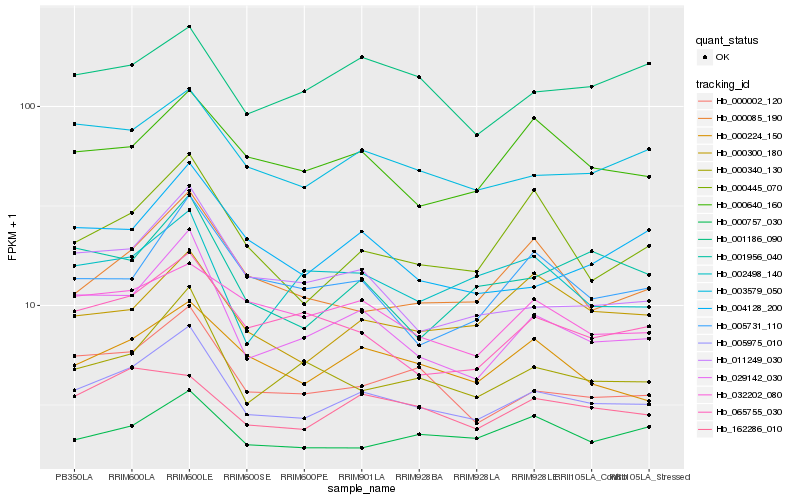
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005975_010 |
0.0 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_029142_030 |
0.0744959626 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 3 |
Hb_003579_050 |
0.0789672318 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 4 |
Hb_000002_120 |
0.07944766 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 5 |
Hb_000340_130 |
0.0859493488 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000224_150 |
0.0937453597 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_065755_030 |
0.0943287505 |
- |
- |
- |
| 8 |
Hb_011249_030 |
0.0995998152 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 9 |
Hb_000640_160 |
0.1005379228 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 10 |
Hb_162286_010 |
0.102156747 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 11 |
Hb_000757_030 |
0.1022622438 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 12 |
Hb_004128_200 |
0.1028866953 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 13 |
Hb_000445_070 |
0.1029838908 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 14 |
Hb_000085_190 |
0.1036287962 |
- |
- |
FtsZ1 protein [Manihot esculenta] |
| 15 |
Hb_000300_180 |
0.1047010318 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 16 |
Hb_032202_080 |
0.1057343952 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001956_040 |
0.1060442272 |
- |
- |
PREDICTED: PGR5-like protein 1B, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001186_090 |
0.1065956444 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 19 |
Hb_005731_110 |
0.1074999886 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 20 |
Hb_002498_140 |
0.1076654566 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |