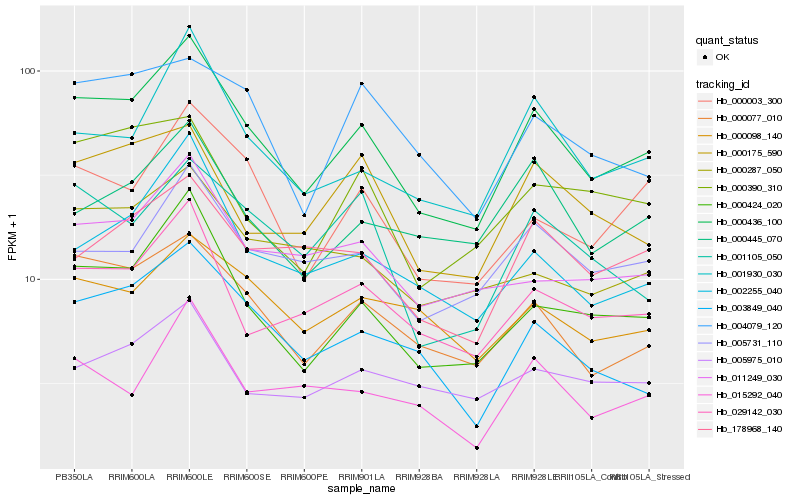
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000436_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 2 |
Hb_000424_020 |
0.0834823061 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 3 |
Hb_000077_010 |
0.0891932333 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_029142_030 |
0.0988106927 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 5 |
Hb_001930_030 |
0.1006327587 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000003_300 |
0.1051848497 |
- |
- |
PREDICTED: uncharacterized protein LOC105631182 [Jatropha curcas] |
| 7 |
Hb_002255_040 |
0.1089997121 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 8 |
Hb_003849_040 |
0.109050313 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 9 |
Hb_005731_110 |
0.1109893032 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 10 |
Hb_011249_030 |
0.1162188926 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 11 |
Hb_178968_140 |
0.1175811009 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000175_590 |
0.1176096472 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000098_140 |
0.1182788133 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 14 |
Hb_000445_070 |
0.1185793456 |
- |
- |
PREDICTED: uncharacterized protein LOC105638998 [Jatropha curcas] |
| 15 |
Hb_000287_050 |
0.119967574 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 16 |
Hb_001105_050 |
0.1205806482 |
- |
- |
PREDICTED: uncharacterized protein LOC105642744 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000390_310 |
0.1207122416 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 18 |
Hb_015292_040 |
0.1211251739 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 19 |
Hb_004079_120 |
0.123284721 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 20 |
Hb_005975_010 |
0.1234706558 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |