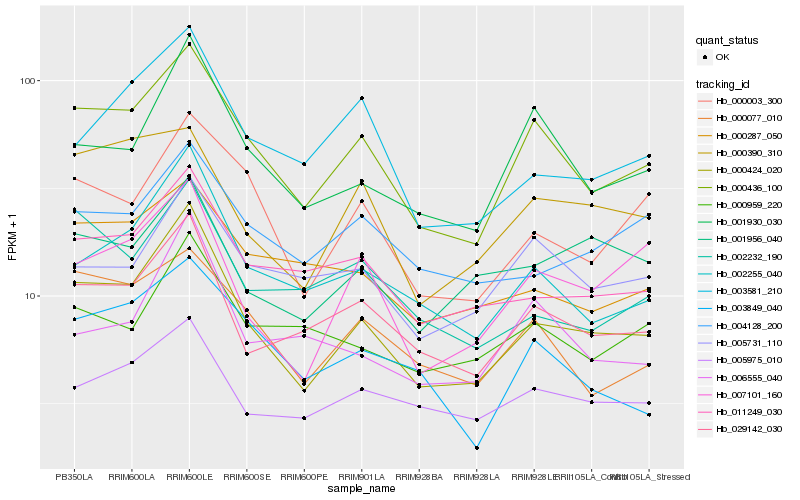
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_020 |
0.0 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 2 |
Hb_000436_100 |
0.0834823061 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 3 |
Hb_029142_030 |
0.1038426853 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 4 |
Hb_002255_040 |
0.1048879352 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 5 |
Hb_011249_030 |
0.1127384694 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 6 |
Hb_001930_030 |
0.1158985555 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000003_300 |
0.1170186982 |
- |
- |
PREDICTED: uncharacterized protein LOC105631182 [Jatropha curcas] |
| 8 |
Hb_007101_160 |
0.1193832601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003581_210 |
0.1257065733 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005975_010 |
0.1281251948 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000390_310 |
0.1292856435 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001956_040 |
0.1299282275 |
- |
- |
PREDICTED: PGR5-like protein 1B, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_004128_200 |
0.1304811168 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 14 |
Hb_000077_010 |
0.1327022135 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_002232_190 |
0.1331004111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000287_050 |
0.13637757 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 17 |
Hb_006555_040 |
0.1371671831 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 18 |
Hb_000959_220 |
0.1397303617 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_003849_040 |
0.1406709537 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 20 |
Hb_005731_110 |
0.1416451791 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |