| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
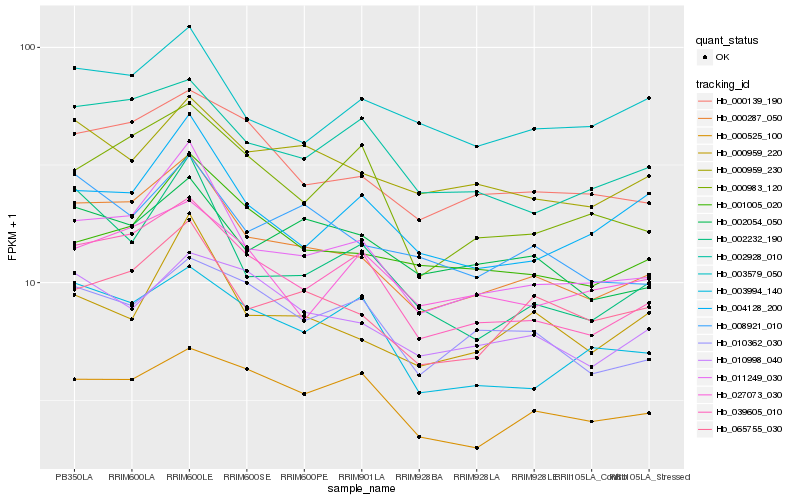
Hb_000287_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 2 |
Hb_011249_030 |
0.0564565556 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 3 |
Hb_039605_010 |
0.0646479734 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000139_190 |
0.0799486247 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_065755_030 |
0.0866539842 |
- |
- |
- |
| 6 |
Hb_002928_010 |
0.0876805669 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 7 |
Hb_008921_010 |
0.0899097338 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 8 |
Hb_002232_190 |
0.09586801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002054_050 |
0.0976879718 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 10 |
Hb_010998_040 |
0.0988156076 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 11 |
Hb_000959_230 |
0.1004935473 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 12 |
Hb_001005_020 |
0.1008814428 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 13 |
Hb_000983_120 |
0.1016983818 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000959_220 |
0.1037598875 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_003994_140 |
0.1040418627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010362_030 |
0.1054719979 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_003579_050 |
0.1056579844 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 18 |
Hb_004128_200 |
0.1073622742 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 19 |
Hb_000525_100 |
0.1075821402 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 20 |
Hb_027073_030 |
0.1082014525 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |