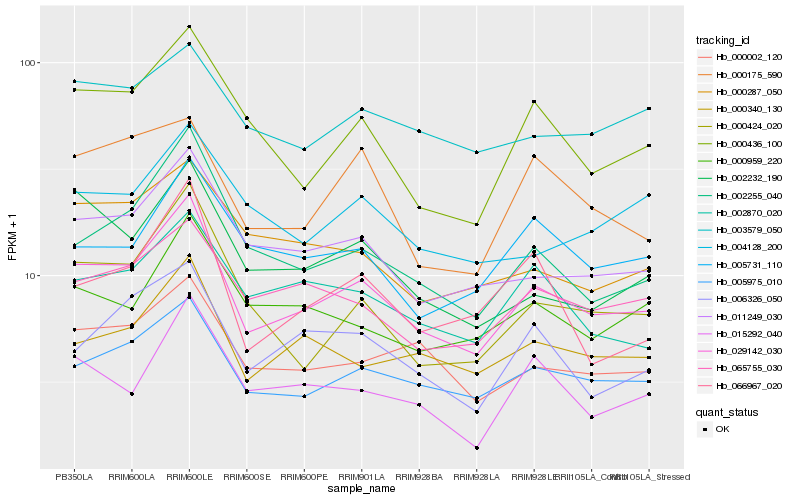
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029142_030 |
0.0 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 2 |
Hb_005975_010 |
0.0744959626 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_011249_030 |
0.0873611279 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 4 |
Hb_000002_120 |
0.0976435987 |
- |
- |
PREDICTED: uncharacterized protein LOC105782064 [Gossypium raimondii] |
| 5 |
Hb_002255_040 |
0.0977666544 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 6 |
Hb_000436_100 |
0.0988106927 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 7 |
Hb_000340_130 |
0.0988332791 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 8 |
Hb_065755_030 |
0.0988703808 |
- |
- |
- |
| 9 |
Hb_000424_020 |
0.1038426853 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 10 |
Hb_002870_020 |
0.1050561408 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002232_190 |
0.1066348524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000959_220 |
0.1074986666 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_005731_110 |
0.1079353817 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 14 |
Hb_006326_050 |
0.1083749758 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_015292_040 |
0.1101164502 |
- |
- |
PREDICTED: myb-like protein X [Jatropha curcas] |
| 16 |
Hb_003579_050 |
0.1108427994 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 17 |
Hb_000287_050 |
0.1125306477 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 18 |
Hb_000175_590 |
0.1143781785 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004128_200 |
0.1159177208 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 20 |
Hb_066967_020 |
0.1166681115 |
- |
- |
RAB6-interacting protein, putative [Ricinus communis] |