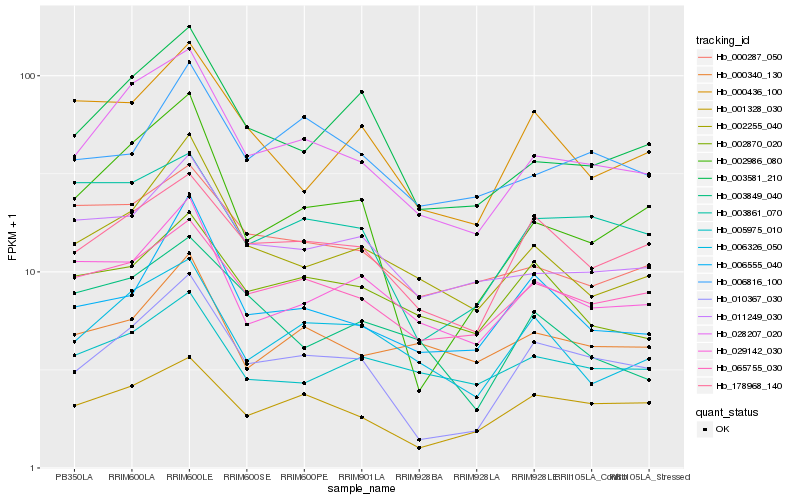
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028207_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650315 isoform X2 [Jatropha curcas] |
| 2 |
Hb_006326_050 |
0.1022278577 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_002255_040 |
0.1040718891 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 4 |
Hb_010367_030 |
0.1049533511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_065755_030 |
0.1054765597 |
- |
- |
- |
| 6 |
Hb_003581_210 |
0.1060992506 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011249_030 |
0.1111890894 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 8 |
Hb_178968_140 |
0.111603701 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_001328_030 |
0.1183486076 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 10 |
Hb_000287_050 |
0.1210881138 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 11 |
Hb_029142_030 |
0.1211066296 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 12 |
Hb_002986_080 |
0.1232018538 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 13 |
Hb_005975_010 |
0.1272065377 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000340_130 |
0.1309441991 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002870_020 |
0.1319492562 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003861_070 |
0.1329169238 |
- |
- |
PREDICTED: uncharacterized protein LOC105650781 [Jatropha curcas] |
| 17 |
Hb_006555_040 |
0.1351761799 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 18 |
Hb_006816_100 |
0.1367435441 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 19 |
Hb_003849_040 |
0.143180011 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 20 |
Hb_000436_100 |
0.1432446187 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |