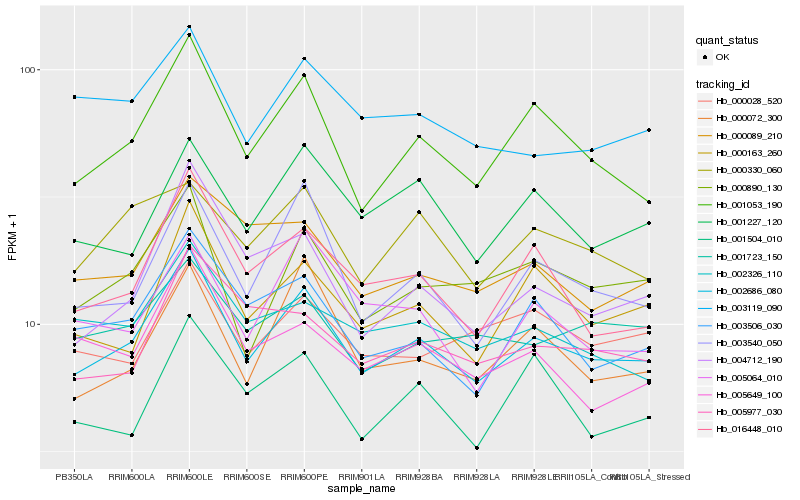
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_080 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 2 |
Hb_001053_190 |
0.0651232801 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 3 |
Hb_003540_050 |
0.0766249703 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 4 |
Hb_000163_260 |
0.0801118512 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 5 |
Hb_000890_130 |
0.0802905415 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 6 |
Hb_016448_010 |
0.081136307 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 7 |
Hb_003506_030 |
0.0817896337 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 8 |
Hb_000072_300 |
0.0853430257 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 9 |
Hb_004712_190 |
0.0857887243 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 10 |
Hb_001227_120 |
0.0858281964 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 11 |
Hb_003119_090 |
0.0905212902 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 12 |
Hb_001723_150 |
0.0906128988 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000330_060 |
0.0907100205 |
- |
- |
PREDICTED: adenosine kinase [Jatropha curcas] |
| 14 |
Hb_002326_110 |
0.0912830908 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 15 |
Hb_000089_210 |
0.0913732126 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_005649_100 |
0.0927007946 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 17 |
Hb_001504_010 |
0.093433835 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 18 |
Hb_000028_520 |
0.094077679 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 19 |
Hb_005977_030 |
0.0943677552 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 20 |
Hb_005064_010 |
0.0952223745 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |