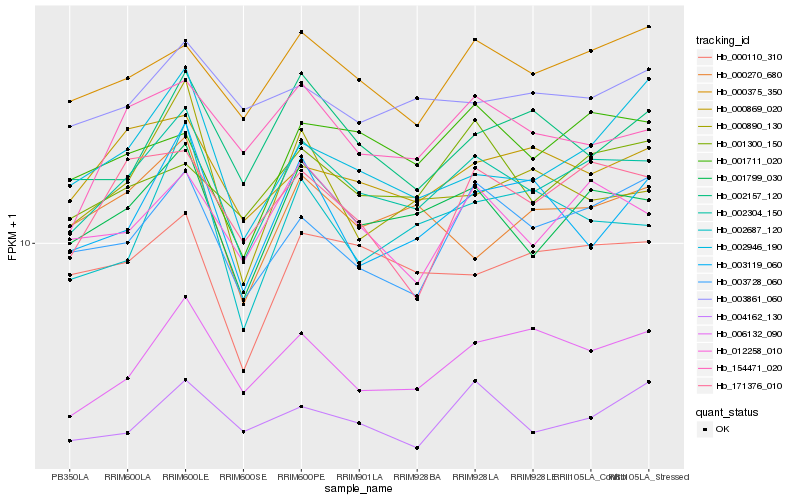
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002304_150 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001799_030 |
0.0479552068 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 3 |
Hb_012258_010 |
0.0758516994 |
- |
- |
PREDICTED: cyclase-associated protein 1 [Jatropha curcas] |
| 4 |
Hb_006132_090 |
0.078248694 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000375_350 |
0.0783282822 |
- |
- |
PREDICTED: uncharacterized protein LOC105641632 [Jatropha curcas] |
| 6 |
Hb_000110_310 |
0.0800075024 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_003728_060 |
0.0823979734 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4-like [Jatropha curcas] |
| 8 |
Hb_154471_020 |
0.0847376385 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |
| 9 |
Hb_000869_020 |
0.0852529848 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 10 |
Hb_171376_010 |
0.0884820413 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001711_020 |
0.0884951097 |
- |
- |
PREDICTED: AP-2 complex subunit mu [Jatropha curcas] |
| 12 |
Hb_002157_120 |
0.090282693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004162_130 |
0.0910866855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003861_060 |
0.0925856407 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 15 |
Hb_000270_680 |
0.0927648543 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 16 |
Hb_002687_120 |
0.0936517005 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000890_130 |
0.093786758 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 18 |
Hb_001300_150 |
0.093792563 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 19 |
Hb_003119_060 |
0.0947420854 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002946_190 |
0.0948976126 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |