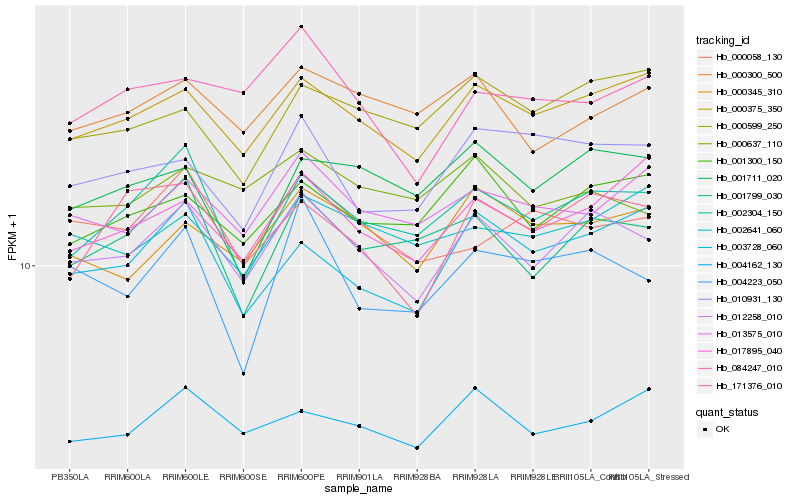
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012258_010 |
0.0 |
- |
- |
PREDICTED: cyclase-associated protein 1 [Jatropha curcas] |
| 2 |
Hb_000375_350 |
0.0681810159 |
- |
- |
PREDICTED: uncharacterized protein LOC105641632 [Jatropha curcas] |
| 3 |
Hb_002304_150 |
0.0758516994 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001799_030 |
0.0826129347 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 5 |
Hb_001300_150 |
0.0863797097 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 6 |
Hb_000345_310 |
0.087188292 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein isoform X3 [Jatropha curcas] |
| 7 |
Hb_171376_010 |
0.0879623872 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 8 |
Hb_013575_010 |
0.0900337213 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 9 |
Hb_000300_500 |
0.0901844783 |
- |
- |
PREDICTED: uncharacterized protein LOC105632604 [Jatropha curcas] |
| 10 |
Hb_000637_110 |
0.092121527 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 11 |
Hb_004162_130 |
0.0931871683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_017895_040 |
0.0940072795 |
- |
- |
hypothetical protein POPTR_0006s08120g [Populus trichocarpa] |
| 13 |
Hb_084247_010 |
0.0959913342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001711_020 |
0.0962217759 |
- |
- |
PREDICTED: AP-2 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_003728_060 |
0.0969998414 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4-like [Jatropha curcas] |
| 16 |
Hb_010931_130 |
0.0979111965 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 17 |
Hb_002641_060 |
0.0982795431 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |
| 18 |
Hb_000599_250 |
0.0983046875 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 19 |
Hb_004223_050 |
0.0985477393 |
- |
- |
hexokinase [Manihot esculenta] |
| 20 |
Hb_000058_130 |
0.0985727117 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |