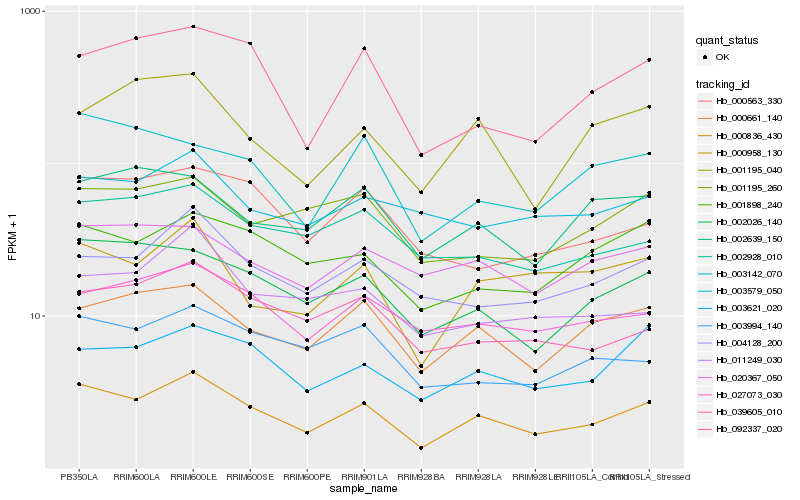
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_430 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_000661_140 |
0.1057140985 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002026_140 |
0.1070146744 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 4 |
Hb_003621_020 |
0.114640481 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 5 |
Hb_004128_200 |
0.1154973707 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 6 |
Hb_000958_130 |
0.117149118 |
- |
- |
PREDICTED: uncharacterized protein LOC105628716 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001898_240 |
0.1182271464 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 8 |
Hb_027073_030 |
0.1187173012 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 9 |
Hb_039605_010 |
0.1209197225 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 10 |
Hb_092337_020 |
0.1223033908 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 11 |
Hb_002928_010 |
0.1234319159 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 12 |
Hb_003994_140 |
0.1240971995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_020367_050 |
0.1251502177 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_000563_330 |
0.1286209063 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_001195_040 |
0.1286941942 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 16 |
Hb_002639_150 |
0.1292175141 |
- |
- |
- |
| 17 |
Hb_003142_070 |
0.1306751294 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 18 |
Hb_003579_050 |
0.1308515086 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 19 |
Hb_001195_260 |
0.1310164611 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 20 |
Hb_011249_030 |
0.1316591629 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |