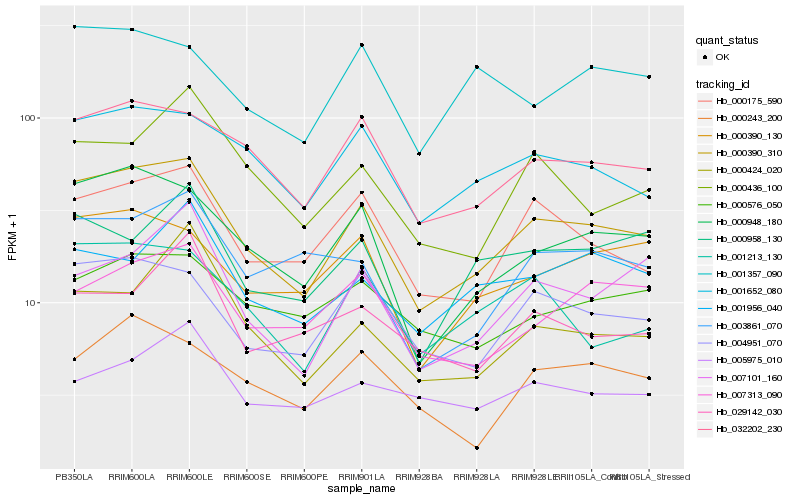
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000948_180 |
0.0850145777 |
- |
- |
PREDICTED: uncharacterized protein LOC105629501 [Jatropha curcas] |
| 3 |
Hb_032202_230 |
0.0909075584 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 4 |
Hb_004951_070 |
0.0928356056 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 5 |
Hb_000175_590 |
0.0970881885 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001652_080 |
0.0980370318 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 7 |
Hb_001213_130 |
0.1110005304 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000390_130 |
0.1120562608 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B isoform X2 [Jatropha curcas] |
| 9 |
Hb_007101_160 |
0.1205589769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000436_100 |
0.1207122416 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 11 |
Hb_007313_090 |
0.1207206494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000243_200 |
0.1215619655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000576_050 |
0.1220331998 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 14 |
Hb_003861_070 |
0.1225306691 |
- |
- |
PREDICTED: uncharacterized protein LOC105650781 [Jatropha curcas] |
| 15 |
Hb_000958_130 |
0.1234205911 |
- |
- |
PREDICTED: uncharacterized protein LOC105628716 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001956_040 |
0.1259482783 |
- |
- |
PREDICTED: PGR5-like protein 1B, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000424_020 |
0.1292856435 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 18 |
Hb_001357_090 |
0.1315247841 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 19 |
Hb_029142_030 |
0.1332247539 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 20 |
Hb_005975_010 |
0.1350167676 |
- |
- |
[ribulose-bisphosphate carboxylase]-lysine N-methyltransferase, putative [Ricinus communis] |