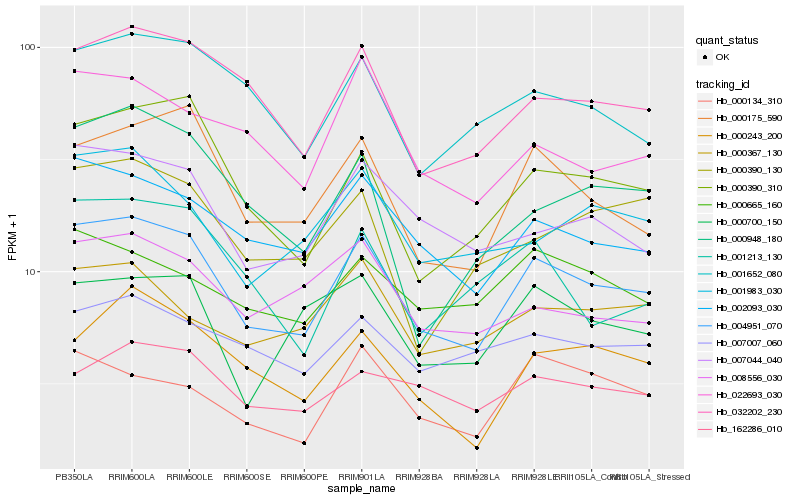
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004951_070 |
0.0 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 2 |
Hb_002093_030 |
0.0870726951 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 3 |
Hb_000175_590 |
0.089162664 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_032202_230 |
0.0924188338 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 5 |
Hb_000390_310 |
0.0928356056 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007044_040 |
0.099012353 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |
| 7 |
Hb_000243_200 |
0.1031371275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001652_080 |
0.1063371006 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 9 |
Hb_001983_030 |
0.1064964314 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |
| 10 |
Hb_022693_030 |
0.1117620916 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 11 |
Hb_008556_030 |
0.112062683 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000367_130 |
0.1136490907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007007_060 |
0.1144438523 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000700_150 |
0.1145579666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001213_130 |
0.114928595 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_162286_010 |
0.1153024479 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 17 |
Hb_000390_130 |
0.1155155591 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B isoform X2 [Jatropha curcas] |
| 18 |
Hb_000948_180 |
0.1162538962 |
- |
- |
PREDICTED: uncharacterized protein LOC105629501 [Jatropha curcas] |
| 19 |
Hb_000665_160 |
0.1166315947 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000134_310 |
0.1169820854 |
- |
- |
PREDICTED: protein gamma response 1 isoform X1 [Jatropha curcas] |