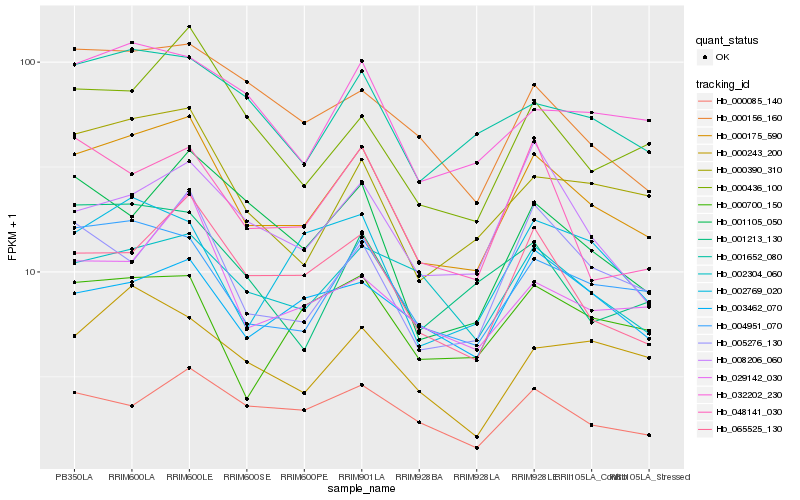
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_590 |
0.0 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004951_070 |
0.089162664 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 3 |
Hb_000390_310 |
0.0970881885 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 4 |
Hb_065525_130 |
0.1044139949 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001652_080 |
0.1105287162 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 6 |
Hb_032202_230 |
0.1130515834 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 7 |
Hb_005276_130 |
0.1139062445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_029142_030 |
0.1143781785 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 9 |
Hb_000436_100 |
0.1176096472 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 10 |
Hb_000700_150 |
0.1209920274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002304_060 |
0.1211607177 |
- |
- |
PREDICTED: glucosidase 2 subunit beta [Jatropha curcas] |
| 12 |
Hb_000085_140 |
0.1211731452 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 13 |
Hb_048141_030 |
0.1219376518 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g59040-like [Prunus mume] |
| 14 |
Hb_000156_160 |
0.1242684582 |
- |
- |
PREDICTED: urease accessory protein G [Jatropha curcas] |
| 15 |
Hb_001213_130 |
0.1260147078 |
- |
- |
PREDICTED: fructokinase-like 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001105_050 |
0.126203635 |
- |
- |
PREDICTED: uncharacterized protein LOC105642744 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008206_060 |
0.1264940559 |
- |
- |
PREDICTED: preprotein translocase subunit SCY1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_002769_020 |
0.1267265622 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 19 |
Hb_000243_200 |
0.1283656371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003462_070 |
0.1285467482 |
- |
- |
arsenical pump-driving atpase, putative [Ricinus communis] |