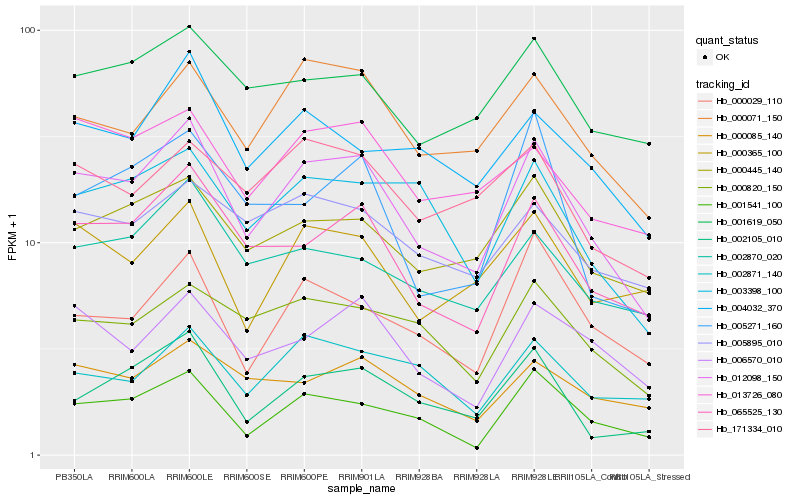
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012098_150 |
0.0 |
- |
- |
ARC5 protein [Manihot esculenta] |
| 2 |
Hb_065525_130 |
0.0901713228 |
- |
- |
PREDICTED: probable methionine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000071_150 |
0.1042374817 |
- |
- |
Stearoy-ACP desaturase [Ricinus communis] |
| 4 |
Hb_001541_100 |
0.1113076737 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 5 |
Hb_000445_140 |
0.1135584454 |
- |
- |
hypothetical protein RCOM_0561550 [Ricinus communis] |
| 6 |
Hb_006570_010 |
0.116006095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003398_100 |
0.1174259507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000820_150 |
0.1189205255 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 9 |
Hb_013726_080 |
0.118959163 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 10 |
Hb_002870_020 |
0.1195667699 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004032_370 |
0.1215271719 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 12 |
Hb_000085_140 |
0.1230576232 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105633025 [Jatropha curcas] |
| 13 |
Hb_002105_010 |
0.1244013261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_171334_010 |
0.1246293088 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 15 |
Hb_005895_010 |
0.1246867482 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001619_050 |
0.1247898851 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000365_100 |
0.1262511732 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000029_110 |
0.1269337835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002871_140 |
0.1284032175 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 20 |
Hb_005271_160 |
0.1308339193 |
- |
- |
PREDICTED: uncharacterized protein LOC105645844 [Jatropha curcas] |