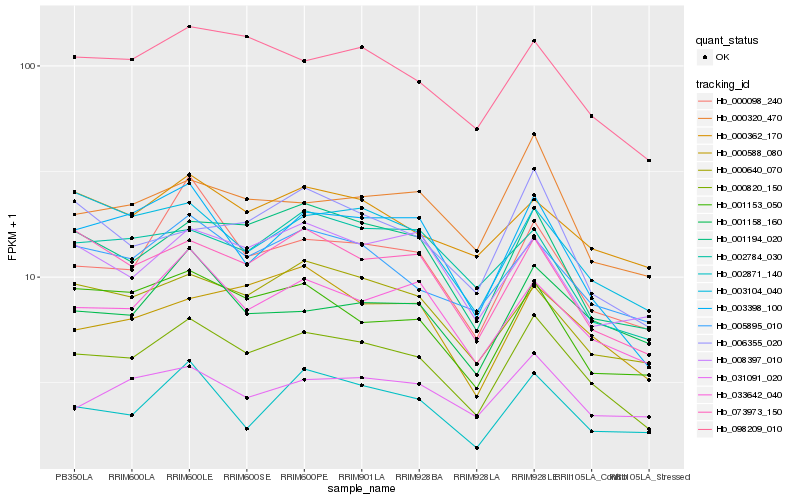
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000820_150 |
0.0 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 2 |
Hb_001194_020 |
0.0787428053 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 3 |
Hb_033642_040 |
0.0793506306 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_098209_010 |
0.080334757 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 5 |
Hb_005895_010 |
0.0805561231 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003398_100 |
0.0807350005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000640_070 |
0.0865381699 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 8 |
Hb_073973_150 |
0.0873972865 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001153_050 |
0.0892526875 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |
| 10 |
Hb_000098_240 |
0.0920348617 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 11 |
Hb_000320_470 |
0.0945950734 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 12 |
Hb_000588_080 |
0.095707941 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 13 |
Hb_003104_040 |
0.0967128064 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 14 |
Hb_000362_170 |
0.0973135678 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 15 |
Hb_008397_010 |
0.0979342854 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001158_160 |
0.0980230161 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002871_140 |
0.0995403799 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 18 |
Hb_031091_020 |
0.1007705773 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 19 |
Hb_002784_030 |
0.1011428281 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 20 |
Hb_006355_020 |
0.1012864428 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |