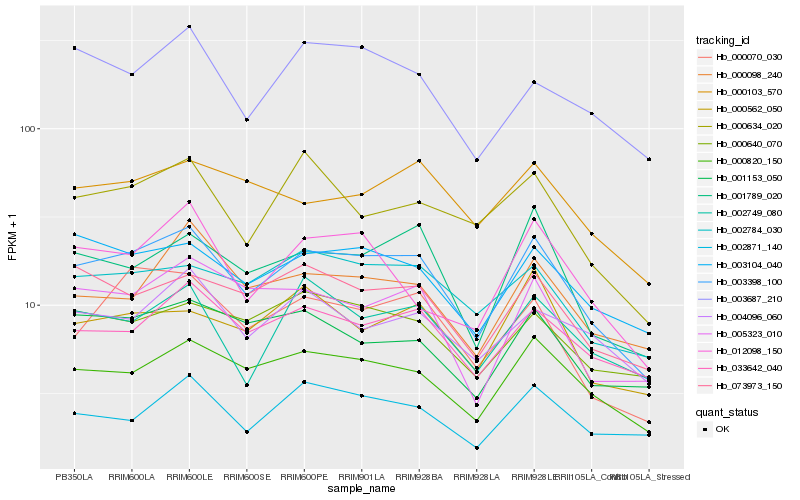
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003398_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000820_150 |
0.0807350005 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 3 |
Hb_033642_040 |
0.0944037032 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003104_040 |
0.0978596579 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 5 |
Hb_001789_020 |
0.1000932965 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002784_030 |
0.1023440455 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 7 |
Hb_073973_150 |
0.1026756113 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000070_030 |
0.1060789065 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 9 |
Hb_005323_010 |
0.1064761963 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 10 |
Hb_001153_050 |
0.1086409104 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |
| 11 |
Hb_000098_240 |
0.1089806955 |
- |
- |
PREDICTED: UDP-glucose:glycoprotein glucosyltransferase [Jatropha curcas] |
| 12 |
Hb_000640_070 |
0.1107330584 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 13 |
Hb_004096_060 |
0.1109020172 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 14 |
Hb_000634_020 |
0.1113398699 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 15 |
Hb_002749_080 |
0.1128713608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002871_140 |
0.1138815425 |
- |
- |
hypothetical protein JCGZ_06847 [Jatropha curcas] |
| 17 |
Hb_000103_570 |
0.113956811 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003687_210 |
0.1144871101 |
- |
- |
PREDICTED: malate dehydrogenase [Populus euphratica] |
| 19 |
Hb_000562_050 |
0.1151223457 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 20 |
Hb_012098_150 |
0.1174259507 |
- |
- |
ARC5 protein [Manihot esculenta] |