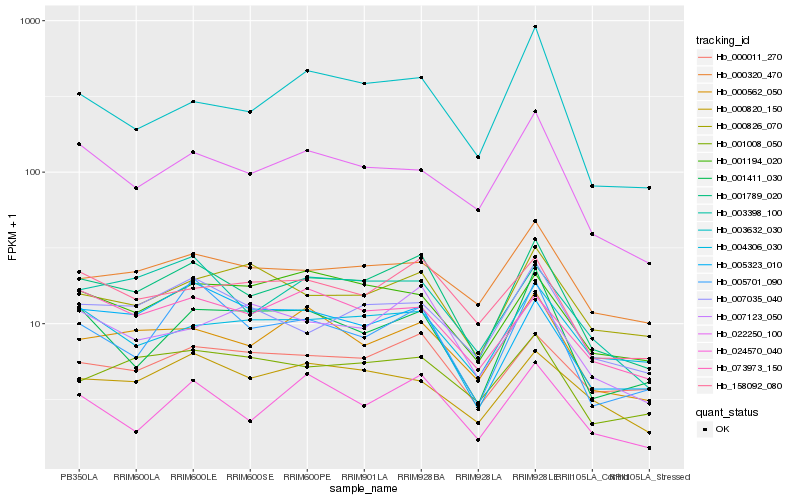
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001789_020 |
0.0 |
- |
- |
PREDICTED: tripeptidyl-peptidase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000562_050 |
0.0955800214 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 3 |
Hb_158092_080 |
0.0997303403 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 4 |
Hb_003398_100 |
0.1000932965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007035_040 |
0.1030086584 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000011_270 |
0.1046555829 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 7 |
Hb_005701_090 |
0.1048861142 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000320_470 |
0.1067429683 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 9 |
Hb_022250_100 |
0.1075842206 |
- |
- |
calnexin, putative [Ricinus communis] |
| 10 |
Hb_001008_050 |
0.1079657211 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 11 |
Hb_024570_040 |
0.1088229619 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 12 |
Hb_005323_010 |
0.1092085592 |
- |
- |
PREDICTED: uncharacterized protein LOC105642810 [Jatropha curcas] |
| 13 |
Hb_007123_050 |
0.1108701012 |
- |
- |
Vitellogenic carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_000820_150 |
0.1118027486 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 15 |
Hb_004306_030 |
0.113787067 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 16 |
Hb_001411_030 |
0.1157079405 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 17 |
Hb_073973_150 |
0.1164807401 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001194_020 |
0.1172166606 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 19 |
Hb_000826_070 |
0.1182153542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003632_030 |
0.1183329836 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |