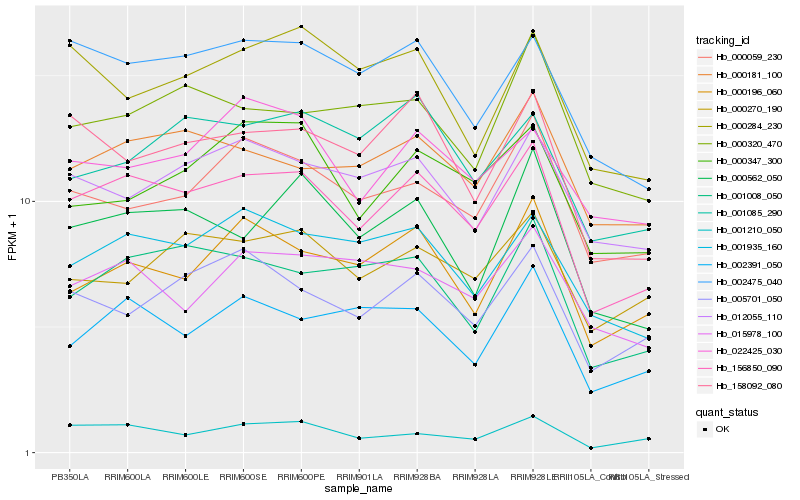
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156850_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 2 |
Hb_002475_040 |
0.0815066068 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001935_160 |
0.0830236354 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 4 |
Hb_001008_050 |
0.0875012657 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_000181_100 |
0.0884075327 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 6 |
Hb_158092_080 |
0.0885849316 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 7 |
Hb_000562_050 |
0.0939027027 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 8 |
Hb_005701_050 |
0.0940945057 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 9 |
Hb_012055_110 |
0.0951209523 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 10 |
Hb_000059_230 |
0.095892183 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 11 |
Hb_001210_050 |
0.095942847 |
- |
- |
hypothetical protein JCGZ_06216 [Jatropha curcas] |
| 12 |
Hb_000196_060 |
0.0963054375 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 13 |
Hb_002391_050 |
0.0976770092 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 14 |
Hb_000270_190 |
0.0992661184 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 15 |
Hb_015978_100 |
0.1029639408 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 16 |
Hb_001085_290 |
0.103197677 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 17 |
Hb_000347_300 |
0.1046861827 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 18 |
Hb_000320_470 |
0.1049335458 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 19 |
Hb_022425_030 |
0.1054819624 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 20 |
Hb_000284_230 |
0.1057226473 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |