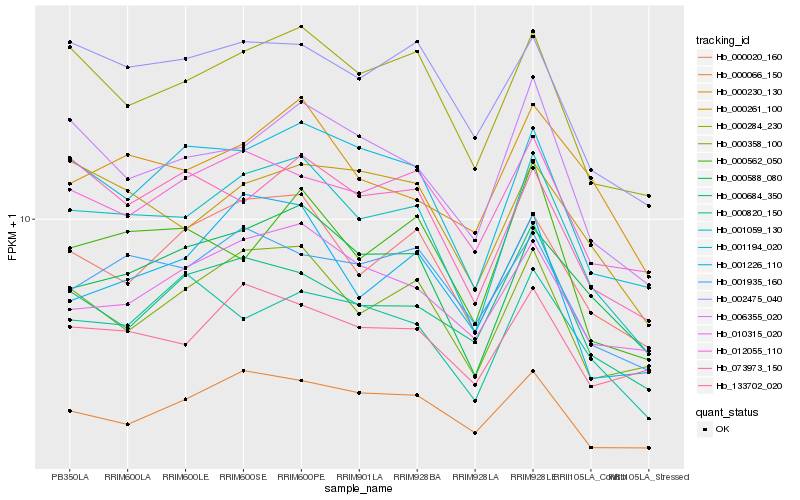
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001059_130 |
0.0 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000284_230 |
0.0750036956 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 3 |
Hb_000588_080 |
0.0770391597 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 4 |
Hb_001194_020 |
0.0776469104 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 5 |
Hb_000358_100 |
0.0780321329 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 6 |
Hb_000020_160 |
0.0800209121 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 7 |
Hb_006355_020 |
0.083746722 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 8 |
Hb_002475_040 |
0.0938201916 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_010315_020 |
0.0966636968 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 10 |
Hb_001935_160 |
0.0966978073 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 11 |
Hb_000261_100 |
0.0968922973 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000066_150 |
0.0971349795 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 13 |
Hb_000562_050 |
0.0974352563 |
- |
- |
PREDICTED: uncharacterized protein LOC105635369 [Jatropha curcas] |
| 14 |
Hb_073973_150 |
0.0985891075 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000684_350 |
0.0996328161 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 16 |
Hb_012055_110 |
0.1004906642 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 17 |
Hb_001226_110 |
0.1012546337 |
- |
- |
PREDICTED: uncharacterized protein LOC105638106 [Jatropha curcas] |
| 18 |
Hb_133702_020 |
0.1012980407 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000230_130 |
0.1024580828 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000820_150 |
0.1048913657 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |