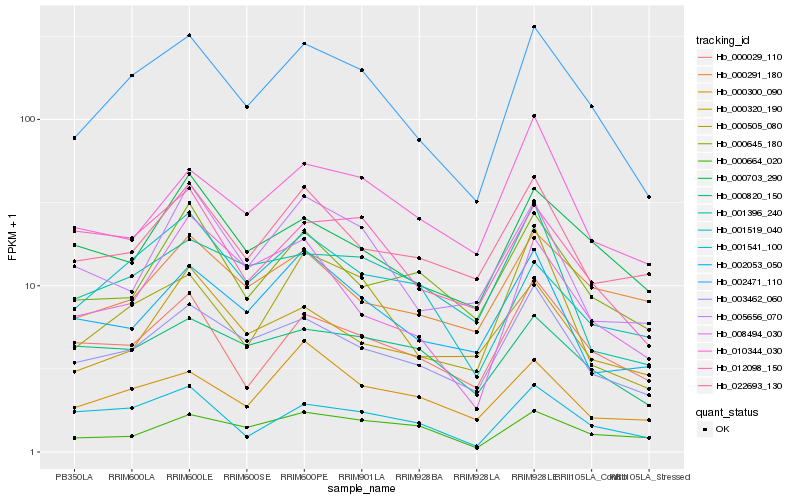
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002471_110 |
0.0 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 2 |
Hb_003462_060 |
0.1067147956 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 3 |
Hb_000505_080 |
0.1290034009 |
- |
- |
Inner membrane transport protein yjjL, putative [Ricinus communis] |
| 4 |
Hb_000029_110 |
0.1315553384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001541_100 |
0.1344405152 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |
| 6 |
Hb_012098_150 |
0.1366351963 |
- |
- |
ARC5 protein [Manihot esculenta] |
| 7 |
Hb_000703_290 |
0.1399996548 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 8 |
Hb_022693_130 |
0.1409598071 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002053_050 |
0.1419844473 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_000300_090 |
0.1434798676 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_001519_040 |
0.1462202176 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 12 |
Hb_000645_180 |
0.1471752182 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 13 |
Hb_000820_150 |
0.1474183964 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 14 |
Hb_000664_020 |
0.1482159372 |
- |
- |
tryptophanyl-tRNA synthetase, putative [Ricinus communis] |
| 15 |
Hb_008494_030 |
0.1486491662 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_000291_180 |
0.1489346966 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 17 |
Hb_001396_240 |
0.1499910415 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_010344_030 |
0.1506431217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005656_070 |
0.1515312984 |
- |
- |
PREDICTED: cryptochrome DASH, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_000320_190 |
0.1516589331 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |