| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
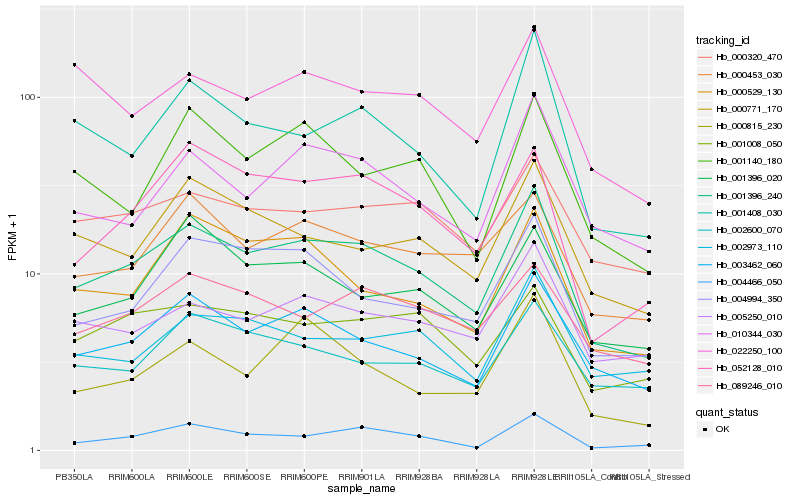
Hb_001396_240 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003462_060 |
0.0892330796 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 3 |
Hb_010344_030 |
0.1063868741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004994_350 |
0.1078969948 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004466_050 |
0.1082018112 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 6 |
Hb_052128_010 |
0.1082943658 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Nicotiana sylvestris] |
| 7 |
Hb_005250_010 |
0.1088620475 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 8 |
Hb_001408_030 |
0.1102552606 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000453_030 |
0.1103157332 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 10 |
Hb_000320_470 |
0.1119742499 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 11 |
Hb_000771_170 |
0.1129830928 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_001008_050 |
0.1148962675 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 13 |
Hb_000529_130 |
0.1153566844 |
- |
- |
hypothetical protein POPTR_0007s14320g [Populus trichocarpa] |
| 14 |
Hb_002973_110 |
0.1162330863 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_002600_070 |
0.1173459328 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 16 |
Hb_089246_010 |
0.1186374982 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 17 |
Hb_001140_180 |
0.1199906284 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 18 |
Hb_001396_020 |
0.1203530919 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 19 |
Hb_022250_100 |
0.1214425828 |
- |
- |
calnexin, putative [Ricinus communis] |
| 20 |
Hb_000815_230 |
0.1224372286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |