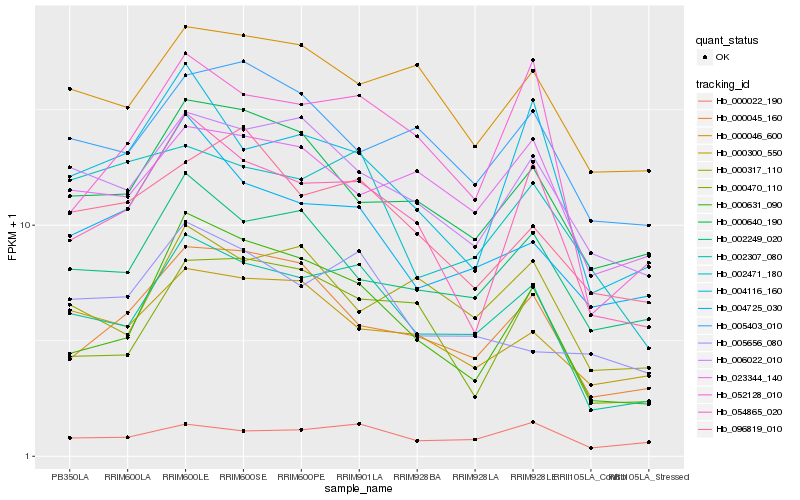
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002307_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006022_010 |
0.1174367153 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 3 |
Hb_054865_020 |
0.117927864 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 4 |
Hb_000631_090 |
0.1202429259 |
- |
- |
PREDICTED: uncharacterized protein LOC105640619 [Jatropha curcas] |
| 5 |
Hb_052128_010 |
0.1226179459 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Nicotiana sylvestris] |
| 6 |
Hb_000045_160 |
0.1328619694 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 7 |
Hb_002249_020 |
0.1333482231 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 8 |
Hb_000046_600 |
0.133769449 |
- |
- |
PREDICTED: cullin-1-like [Jatropha curcas] |
| 9 |
Hb_000640_190 |
0.1338436335 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000022_190 |
0.1348814867 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 11 |
Hb_023344_140 |
0.1366942545 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 12 |
Hb_000470_110 |
0.1375585427 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |
| 13 |
Hb_000300_550 |
0.1377300179 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_004725_030 |
0.1381314538 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 15 |
Hb_004116_160 |
0.1385853663 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005403_010 |
0.1393100925 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 17 |
Hb_000317_110 |
0.1401654979 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B1, mitochondrial isoform X2 [Jatropha curcas] |
| 18 |
Hb_096819_010 |
0.1409613286 |
- |
- |
PREDICTED: GRIP and coiled-coil domain-containing protein 2 isoform X4 [Jatropha curcas] |
| 19 |
Hb_002471_180 |
0.1412559586 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_005656_080 |
0.1414390505 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |