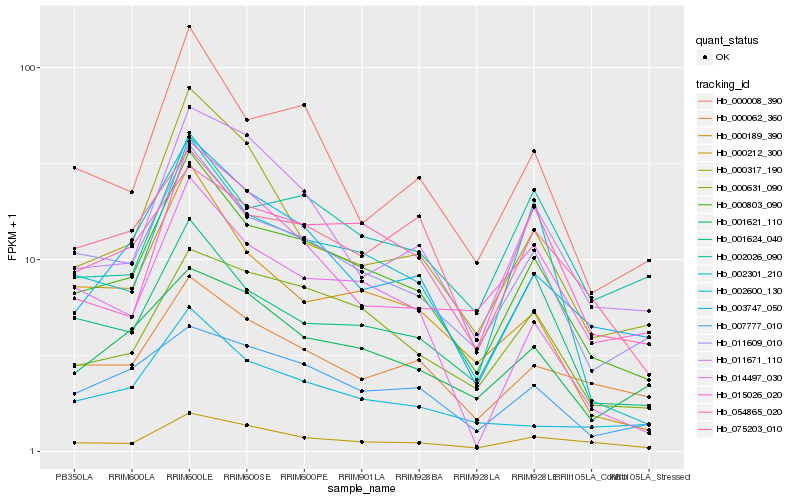
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_090 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 2 |
Hb_011609_010 |
0.0741756477 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 3 |
Hb_003747_050 |
0.1075591998 |
- |
- |
PREDICTED: WD repeat-containing protein LWD1 [Jatropha curcas] |
| 4 |
Hb_000008_390 |
0.1109534954 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 5 |
Hb_001624_040 |
0.1139282255 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002301_210 |
0.1200276274 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000631_090 |
0.122145785 |
- |
- |
PREDICTED: uncharacterized protein LOC105640619 [Jatropha curcas] |
| 8 |
Hb_000212_300 |
0.1228490843 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 9 |
Hb_054865_020 |
0.1278669109 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 10 |
Hb_000189_390 |
0.1284513472 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000062_360 |
0.1287356252 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 12 |
Hb_075203_010 |
0.1304018452 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007777_010 |
0.1323242623 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 14 |
Hb_014497_030 |
0.1323839716 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 15 |
Hb_011671_110 |
0.1328917617 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 16 |
Hb_001621_110 |
0.133128612 |
- |
- |
PREDICTED: uncharacterized protein LOC105647362 [Jatropha curcas] |
| 17 |
Hb_002026_090 |
0.1374656517 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |
| 18 |
Hb_015026_020 |
0.1384941335 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 19 |
Hb_000317_190 |
0.1390494657 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 20 |
Hb_002600_130 |
0.1391685773 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |