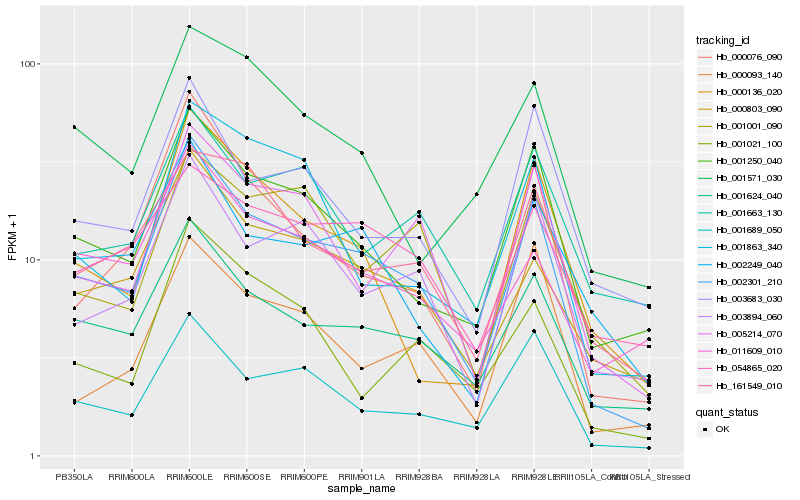
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_210 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001624_040 |
0.1044593796 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001250_040 |
0.1169771389 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 4 |
Hb_000803_090 |
0.1200276274 |
- |
- |
PREDICTED: transcription initiation factor IIB isoform X1 [Jatropha curcas] |
| 5 |
Hb_001689_050 |
0.125082651 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 6 |
Hb_000076_090 |
0.1272043627 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_003894_060 |
0.129822253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005214_070 |
0.1305847724 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 9 |
Hb_002249_040 |
0.138366242 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1-like [Jatropha curcas] |
| 10 |
Hb_003683_030 |
0.1417159503 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 11 |
Hb_011609_010 |
0.1457410378 |
- |
- |
PREDICTED: zinc transporter 5 [Jatropha curcas] |
| 12 |
Hb_161549_010 |
0.1470522795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000136_020 |
0.1474370811 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g80440-like [Jatropha curcas] |
| 14 |
Hb_001021_100 |
0.1495549702 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 15 |
Hb_001663_130 |
0.1558723656 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 16 |
Hb_001001_090 |
0.1564421314 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 17 |
Hb_001863_340 |
0.1585030289 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 18 |
Hb_054865_020 |
0.1586807792 |
transcription factor |
TF Family: HB |
homeobox leucine zipper family protein [Populus trichocarpa] |
| 19 |
Hb_000093_140 |
0.1599970522 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001571_030 |
0.1606967201 |
- |
- |
PREDICTED: uncharacterized protein LOC105640040 isoform X1 [Jatropha curcas] |