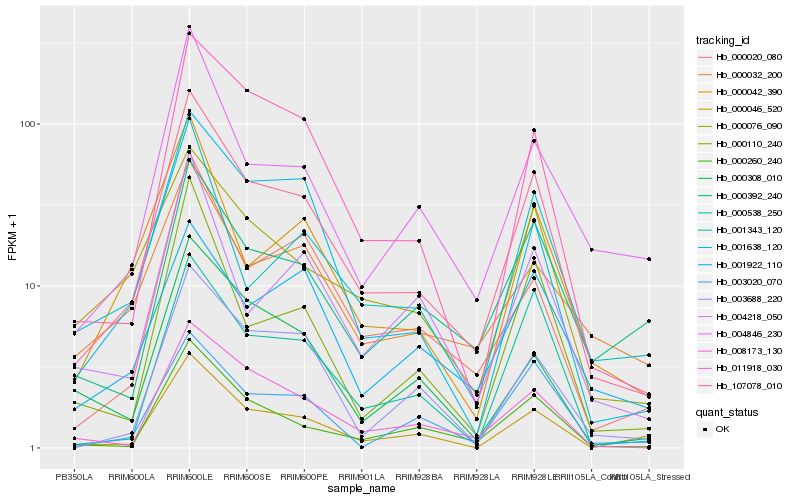
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107078_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_011918_030 |
0.1246320746 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 3 |
Hb_008173_130 |
0.1272719229 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000260_240 |
0.1331977571 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 5 |
Hb_001638_120 |
0.1360889782 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 6 |
Hb_000020_080 |
0.1380368553 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 7 |
Hb_000076_090 |
0.1449907362 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000538_250 |
0.1451351633 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 [Jatropha curcas] |
| 9 |
Hb_000042_390 |
0.148306746 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 10 |
Hb_000046_520 |
0.1495056752 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003688_220 |
0.1504509765 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_004846_230 |
0.1546216054 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000110_240 |
0.1602663264 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000392_240 |
0.1612024842 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001922_110 |
0.164292204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003020_070 |
0.1648623779 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 17 |
Hb_004218_050 |
0.165653421 |
- |
- |
annexin [Manihot esculenta] |
| 18 |
Hb_001343_120 |
0.1670696825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000308_010 |
0.1674870001 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 20 |
Hb_000032_200 |
0.1685121548 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |