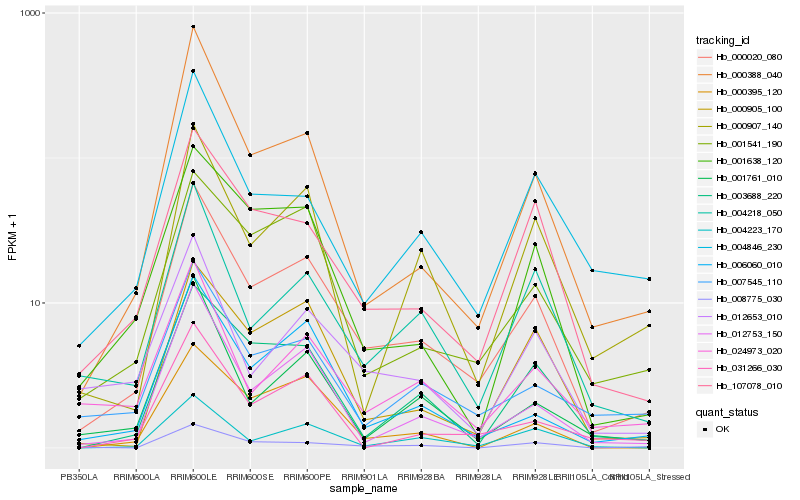
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000020_080 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 2 |
Hb_012753_150 |
0.1332060174 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 3 |
Hb_107078_010 |
0.1380368553 |
- |
- |
- |
| 4 |
Hb_000388_040 |
0.1449572311 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 5 |
Hb_006060_010 |
0.1482851412 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 6 |
Hb_004218_050 |
0.1508366247 |
- |
- |
annexin [Manihot esculenta] |
| 7 |
Hb_007545_110 |
0.1526945095 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_003688_220 |
0.1538184299 |
- |
- |
unknown [Populus trichocarpa] |
| 9 |
Hb_001638_120 |
0.1553425834 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 10 |
Hb_001541_190 |
0.1564402504 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 11 |
Hb_000905_100 |
0.1579810782 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000907_140 |
0.1624694601 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 13 |
Hb_004223_170 |
0.1625659875 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 14 |
Hb_000395_120 |
0.1638485251 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 15 |
Hb_001761_010 |
0.1639916774 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 16 |
Hb_012653_010 |
0.164098945 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 17 |
Hb_024973_020 |
0.1655634131 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 18 |
Hb_004846_230 |
0.1671791676 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_008775_030 |
0.1695877899 |
- |
- |
PREDICTED: ferric reduction oxidase 7, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_031266_030 |
0.1698836577 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |