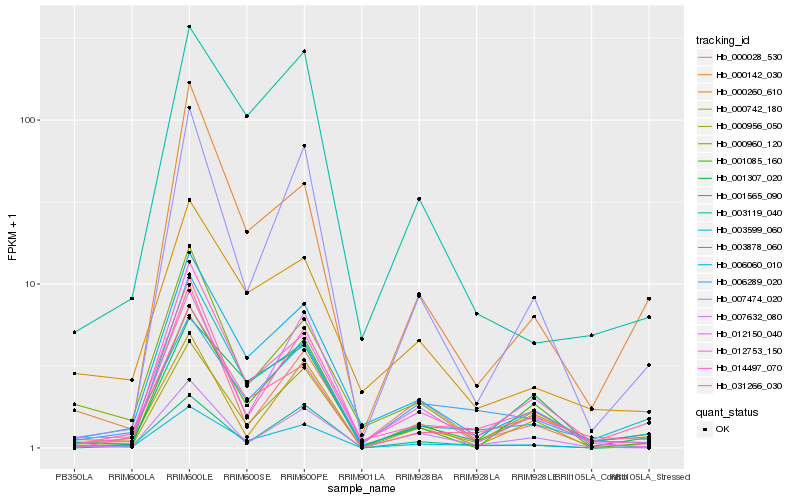
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003878_060 |
0.0 |
- |
- |
hypothetical protein B456_007G326900, partial [Gossypium raimondii] |
| 2 |
Hb_006060_010 |
0.1289026267 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 3 |
Hb_003599_060 |
0.1339771109 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_014497_070 |
0.1397940435 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 5 |
Hb_006289_020 |
0.1451560157 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_012753_150 |
0.1463227257 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 7 |
Hb_007474_020 |
0.1473096612 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 8 |
Hb_031266_030 |
0.1494618473 |
- |
- |
Kinase superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_001565_090 |
0.1535217075 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000960_120 |
0.1610808711 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 11 |
Hb_012150_040 |
0.1616629665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000956_050 |
0.1639676763 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000028_530 |
0.1649766621 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 14 |
Hb_001085_160 |
0.1654049118 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 15 |
Hb_000742_180 |
0.1657788904 |
- |
- |
hypothetical protein POPTR_0011s16640g [Populus trichocarpa] |
| 16 |
Hb_003119_040 |
0.1698245654 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 17 |
Hb_001307_020 |
0.1715593169 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 18 |
Hb_000142_030 |
0.1738026799 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_007632_080 |
0.1740073584 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 20 |
Hb_000260_610 |
0.1743073582 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |